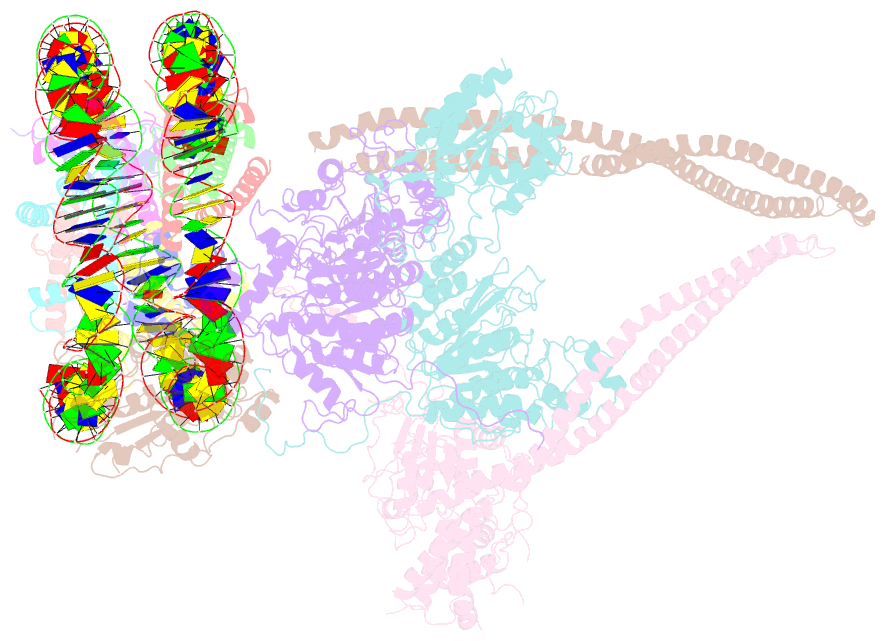
Summary information and primary citation
- PDB-id
- 6z6p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (4.43 Å)
- Summary
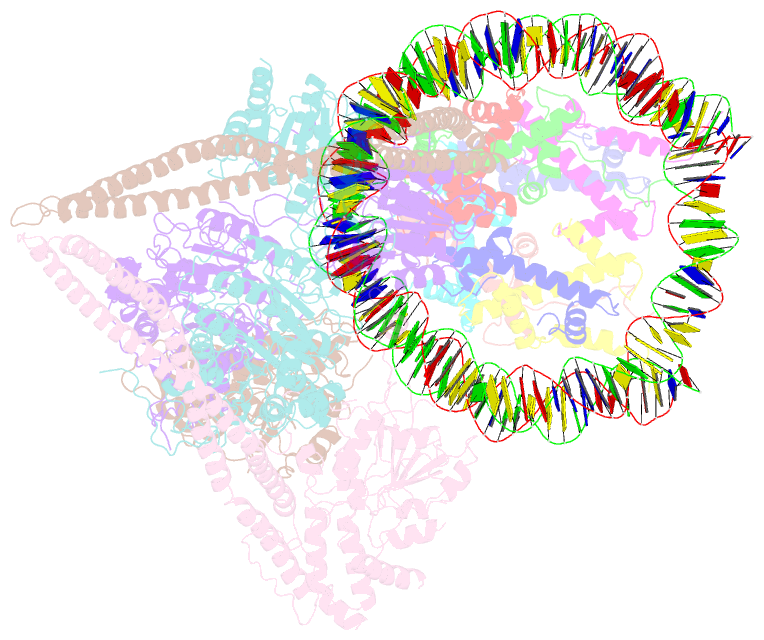
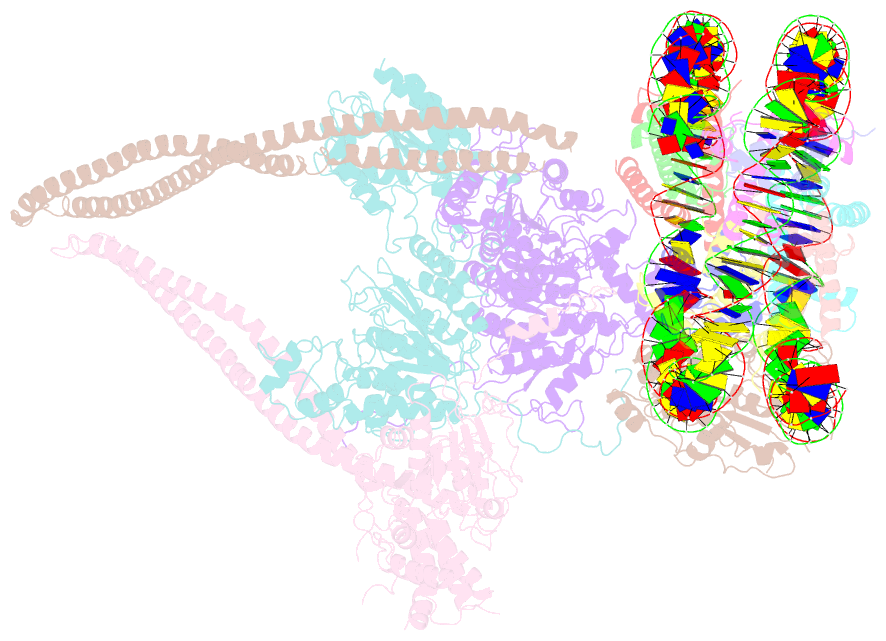
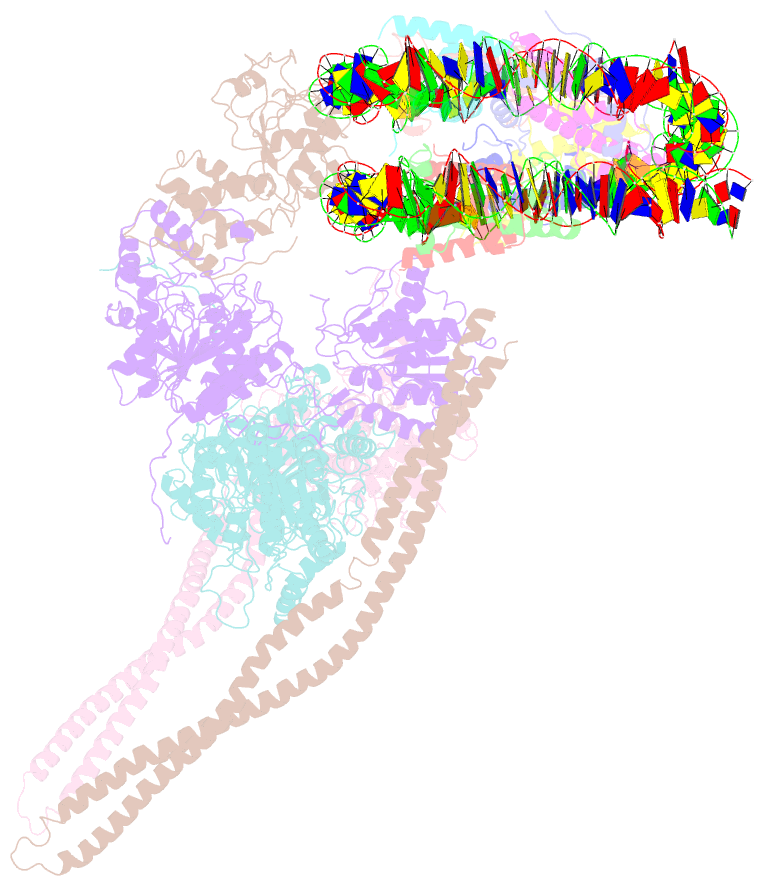
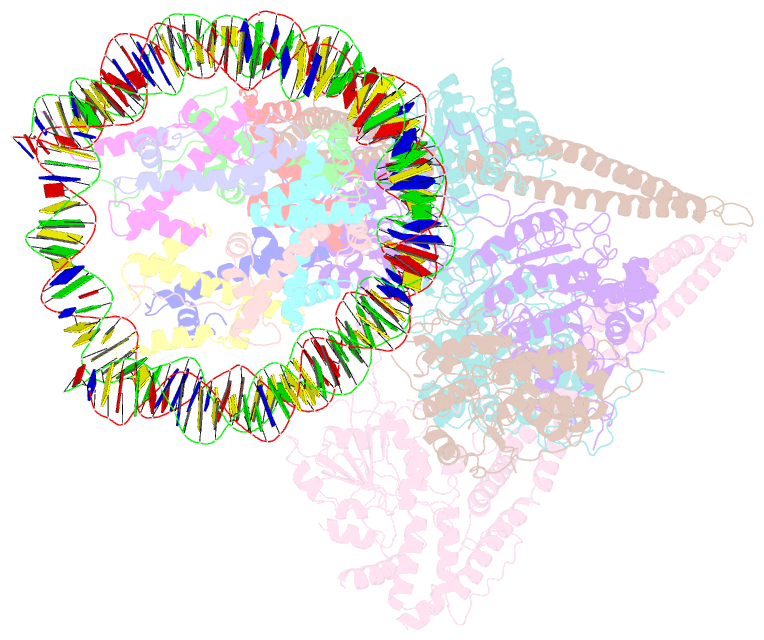
- Hdac-pc-nuc
- Reference
- Lee JH, Bollschweiler D, Schafer T, Huber R (2021): "Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies." Sci Adv, 7. doi: 10.1126/sciadv.abd4413.
- Abstract
- The chromatin-modifying histone deacetylases (HDACs) remove acetyl groups from acetyl-lysine residues in histone amino-terminal tails, thereby mediating transcriptional repression. Structural makeup and mechanisms by which multisubunit HDAC complexes recognize nucleosomes remain elusive. Our cryo-electron microscopy structures of the yeast class II HDAC ensembles show that the HDAC protomer comprises a triangle-shaped assembly of stoichiometry Hda12-Hda2-Hda3, in which the active sites of the Hda1 dimer are freely accessible. We also observe a tetramer of protomers, where the nucleosome binding modules are inaccessible. Structural analysis of the nucleosome-bound complexes indicates how positioning of Hda1 adjacent to histone H2B affords HDAC catalysis. Moreover, it reveals how an intricate network of multiple contacts between a dimer of protomers and the nucleosome creates a platform for expansion of the HDAC activities. Our study provides comprehensive insight into the structural plasticity of the HDAC complex and its functional mechanism of chromatin modification.