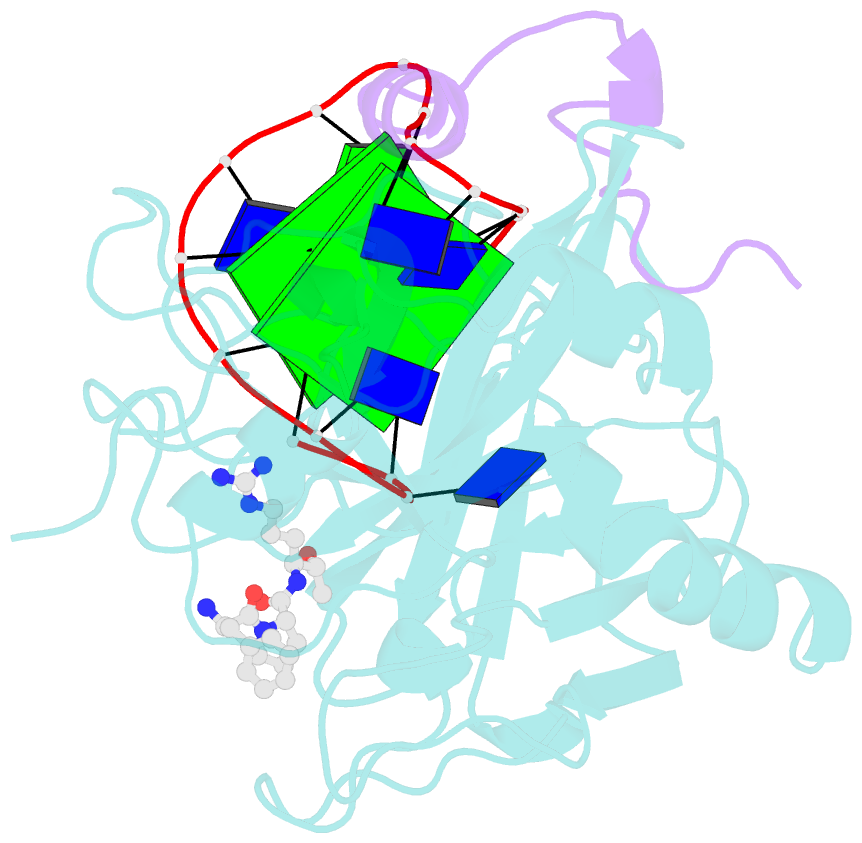
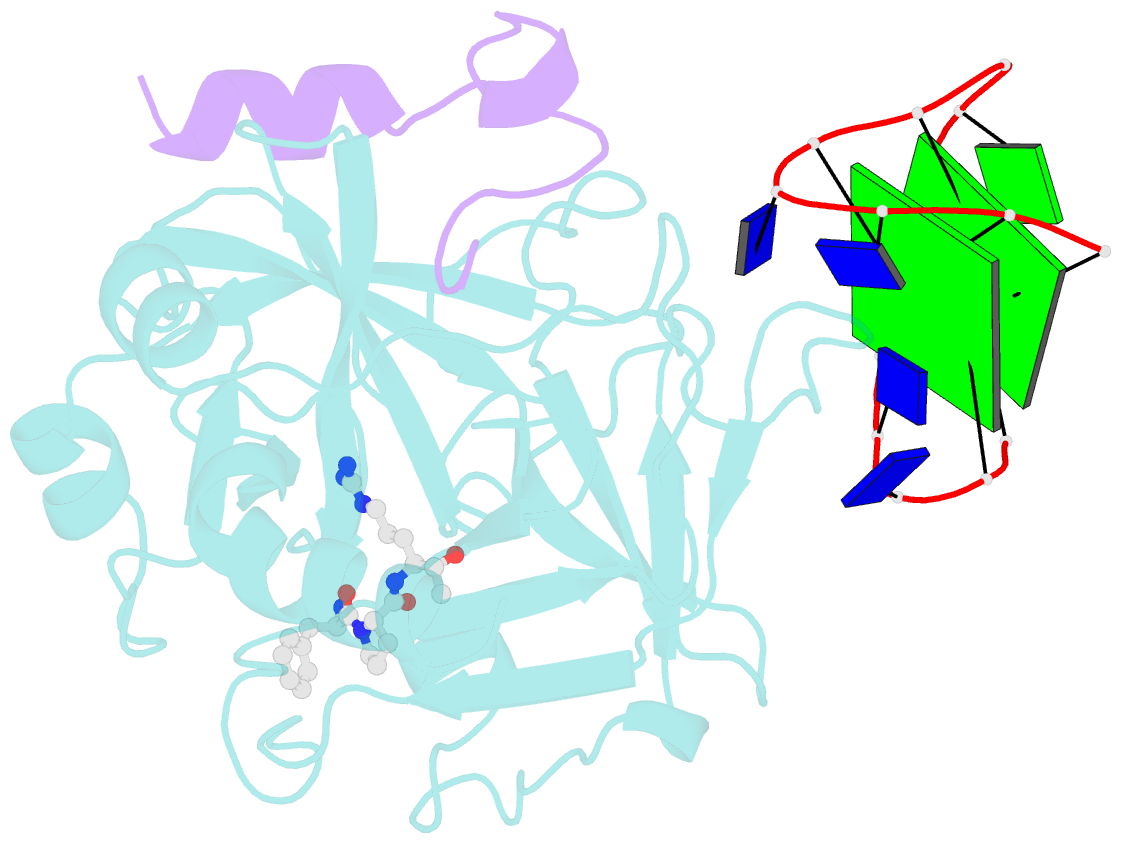
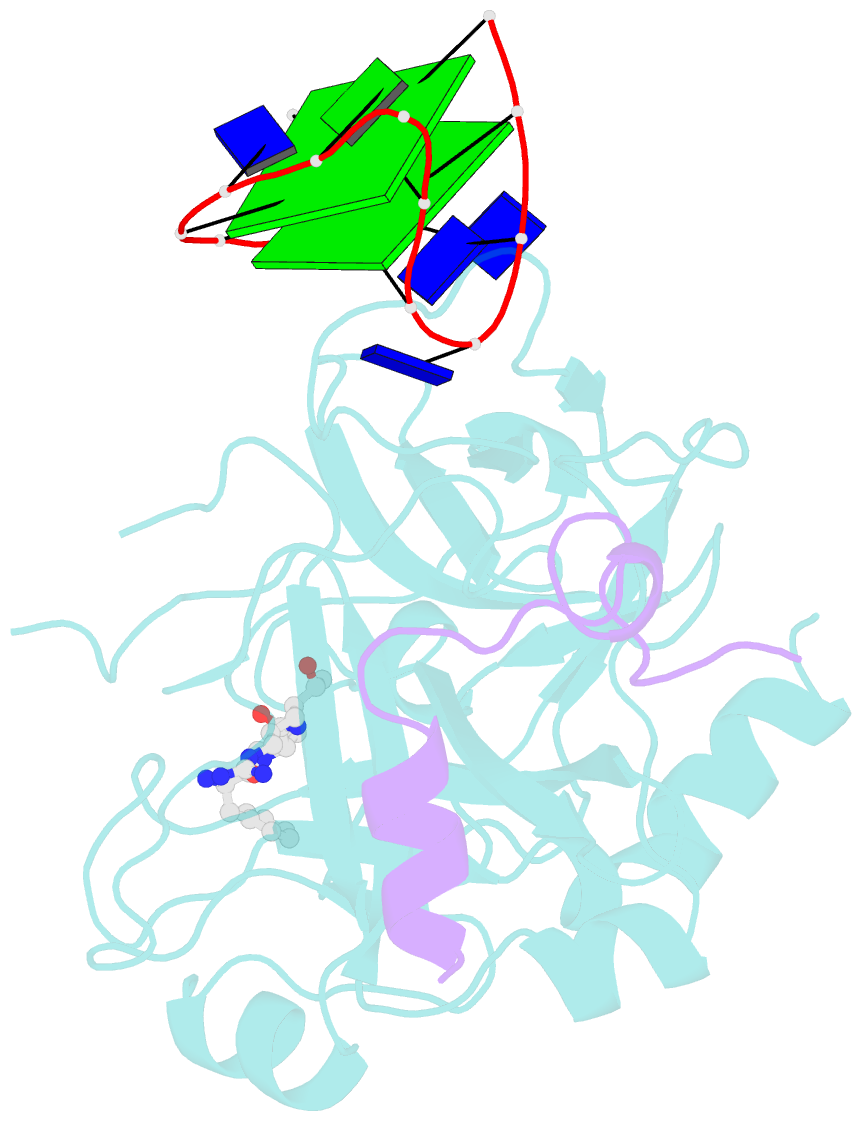
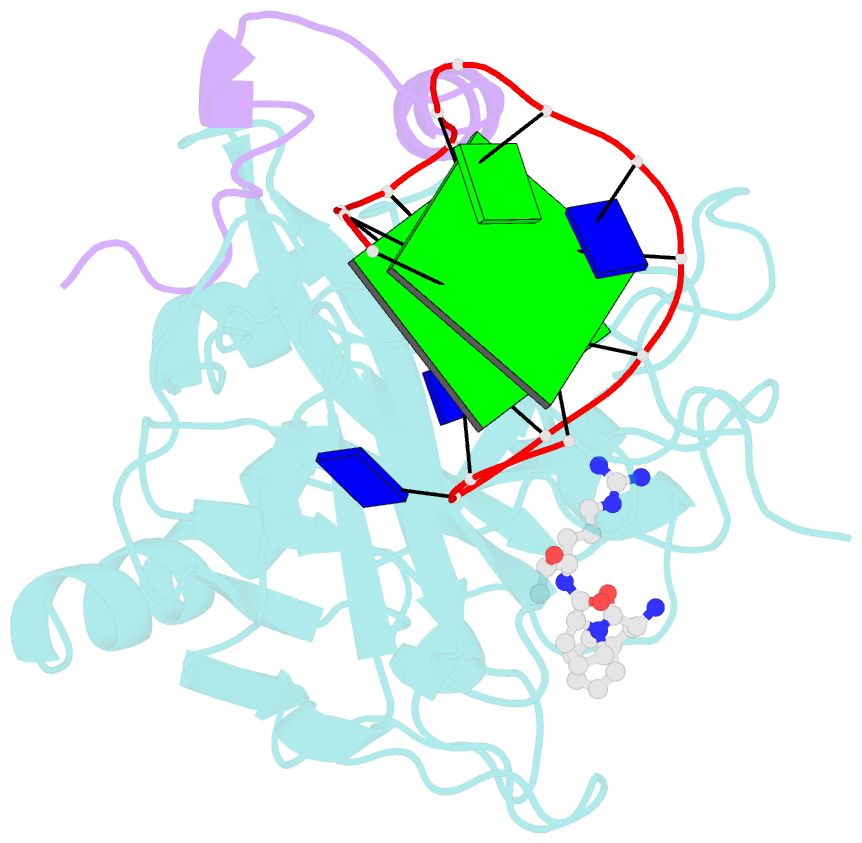
Summary information and primary citation
- PDB-id
- 6z8v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (1.58 Å)
- Summary
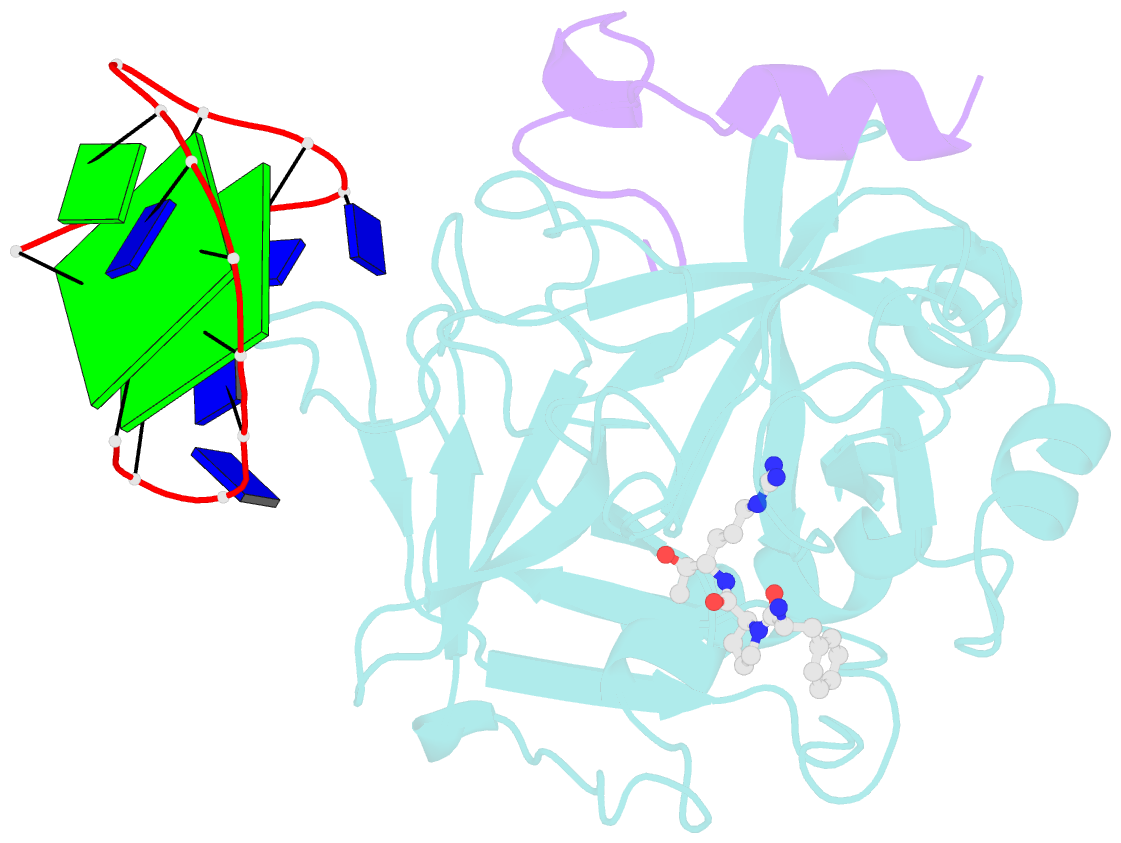
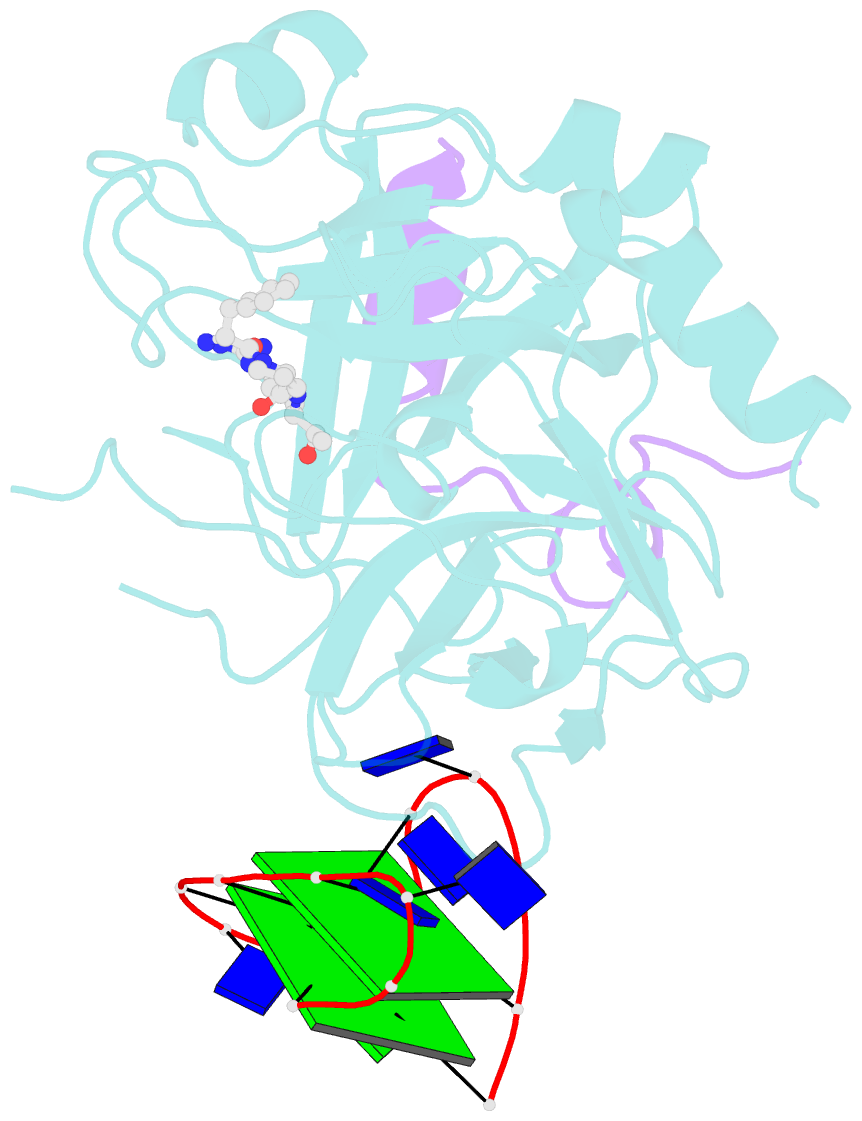
- X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (tba-3l), which contains 1-beta-d-lactopyranosyl residue in the side chain of thy3 at n3.
- Reference
- Smirnov I, Kolganova N, Troisi R, Sica F, Timofeev E (2021): "Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12." Mol Ther Nucleic Acids, 23, 863-871. doi: 10.1016/j.omtn.2021.01.004.
- Abstract
- Post-SELEX modification of DNA aptamers is an established strategy to improve their affinity or inhibitory characteristics. In this study, we examined the possibility of increasing the recognition interface between the thrombin-binding aptamer HD1 (TBA) and thrombin by adding a chemically modified side chain to selected nucleotide residues. A panel of 22 TBA variants with N3-modified residues T3 and T12 was prepared by a two-step modification procedure. Aptamers were characterized by a combination of biophysical and biochemical methods. We identified mutants with enhanced affinity and improved anticoagulant activity. The crystal structures of thrombin complexes with three selected modified variants revealed that the modified pyrimidine base invariably allocates in proximity to thrombin residues Tyr76 and Ile82 due to the directing role of the unmodified TT loop. The modifications induced an increase in the contact areas between thrombin and the modified TBAs. Comparative analysis of the structural, biochemical, and biophysical data suggests that the non-equivalent binding modes of the mutants with thrombin in the T3- and T12-modified series account for the observed systematic differences in their affinity characteristics. In this study, we show that extending the recognition surface between the protein and modified aptamers is a promising approach that may improve characteristics of aptamer ligands.