Summary information and primary citation
- PDB-id
- 6zhe; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (7.24 Å)
- Summary
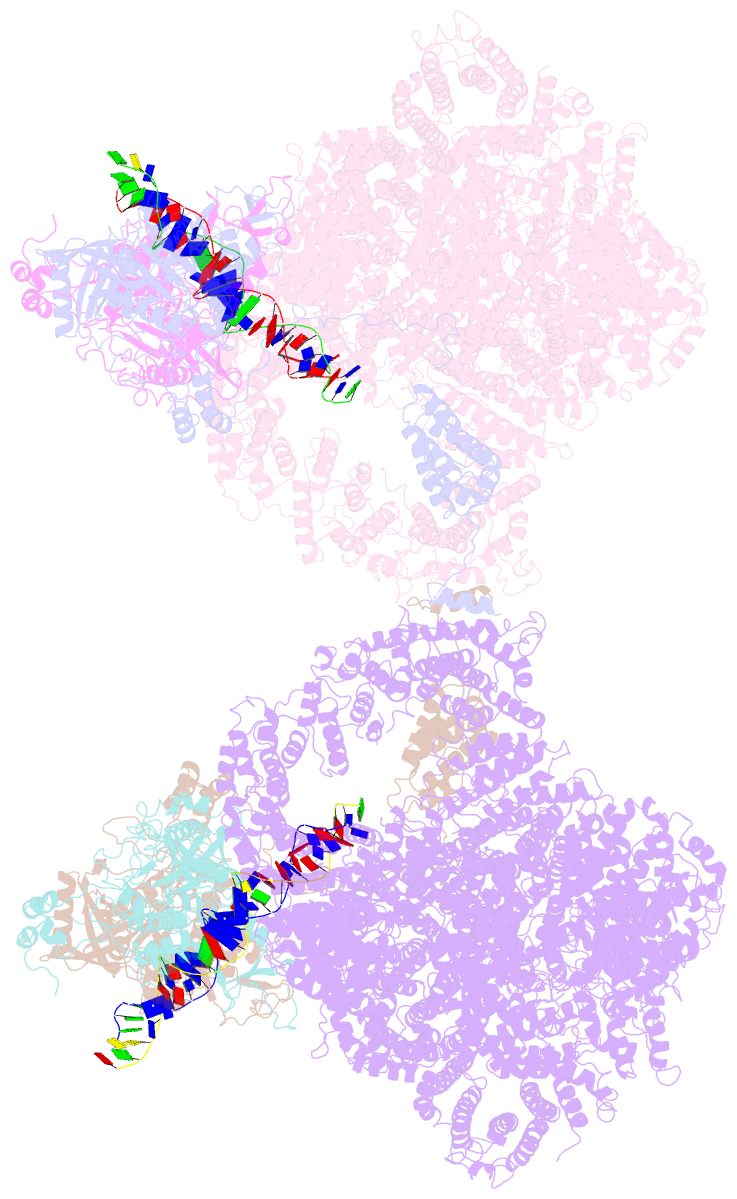
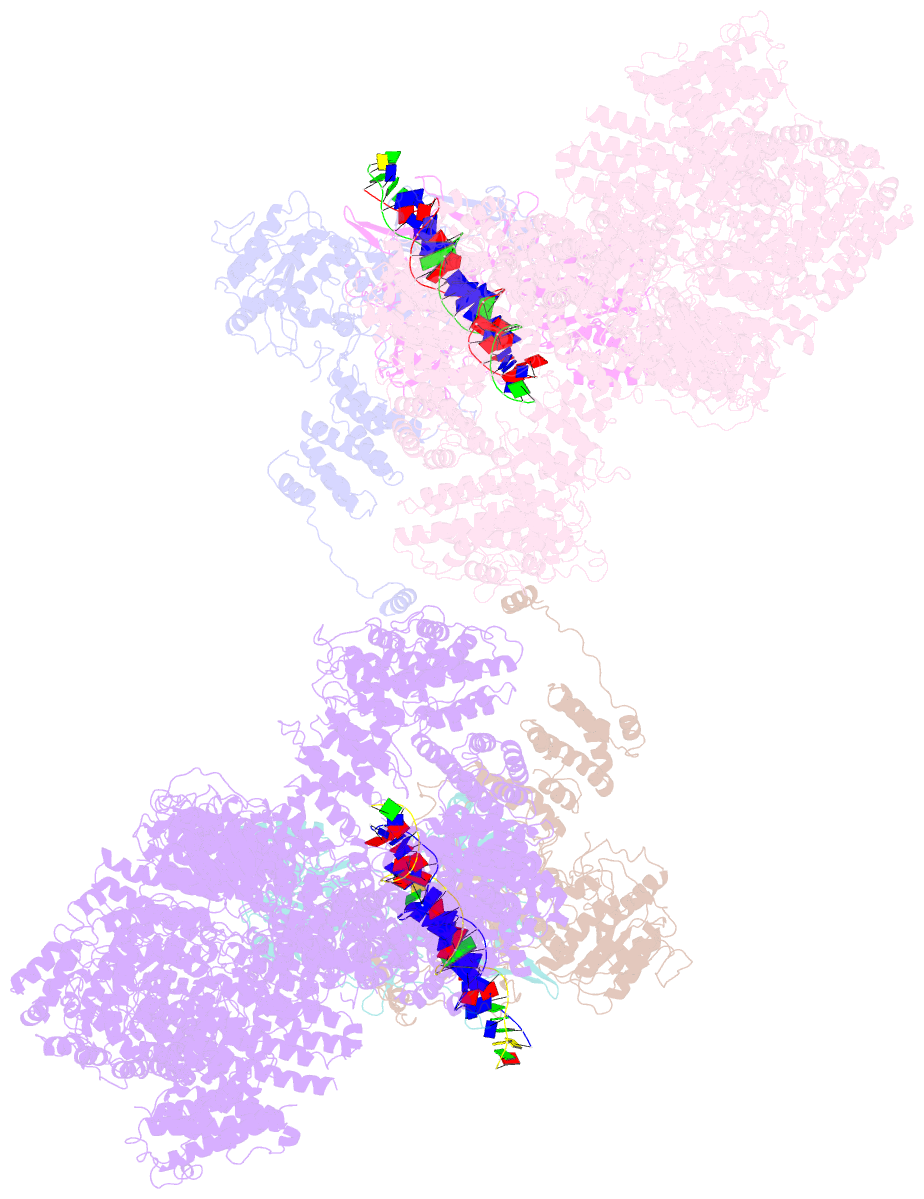
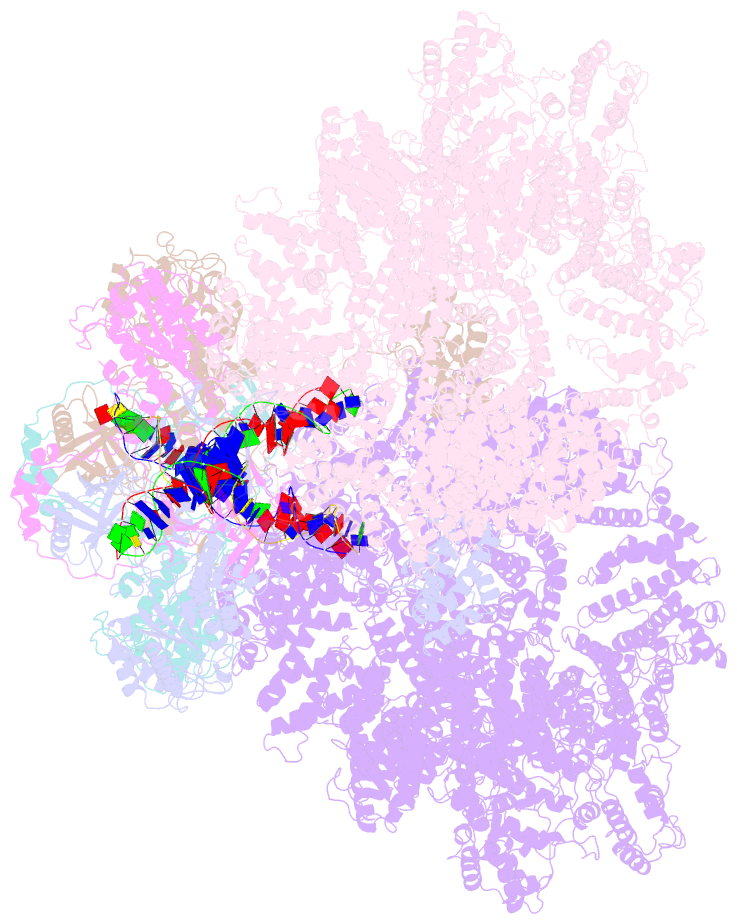
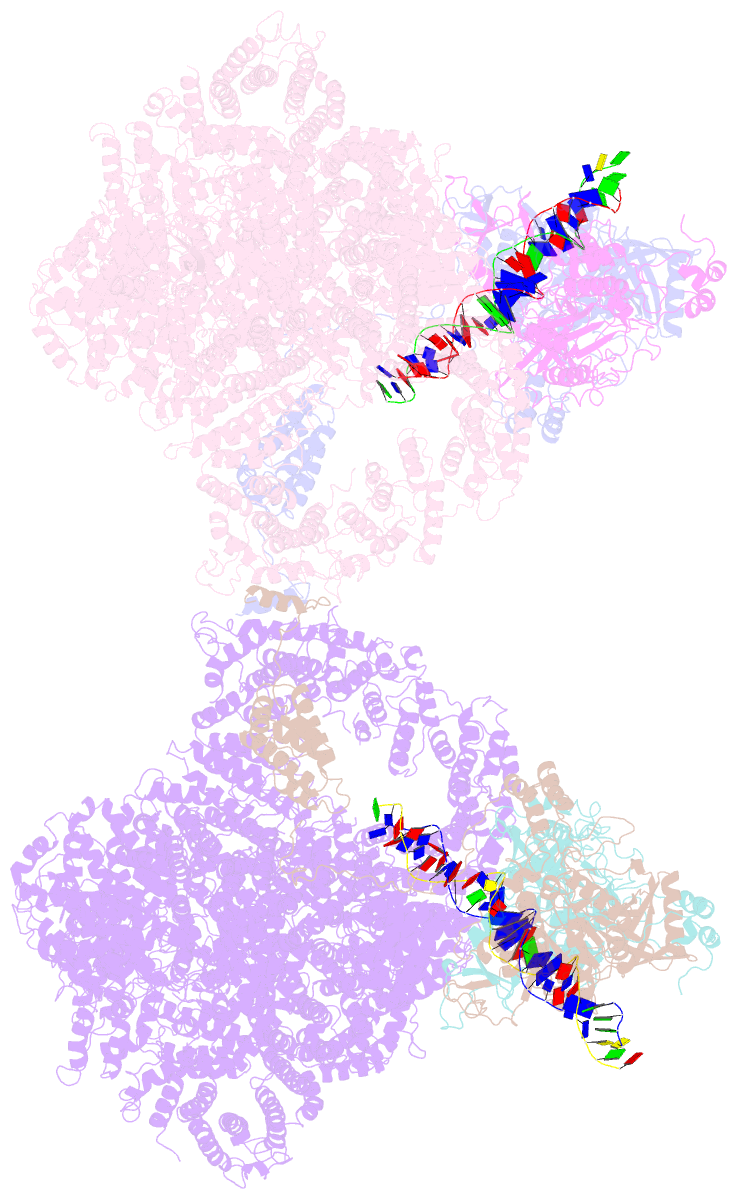
- cryo-EM structure of DNA-pk dimer
- Reference
- Chaplin AK, Hardwick SW, Liang S, Kefala Stavridi A, Hnizda A, Cooper LR, De Oliveira TM, Chirgadze DY, Blundell TL (2021): "Dimers of DNA-PK create a stage for DNA double-strand break repair." Nat.Struct.Mol.Biol., 28, 13-19. doi: 10.1038/s41594-020-00517-x.
- Abstract
- DNA double-strand breaks are the most dangerous type of DNA damage and, if not repaired correctly, can lead to cancer. In humans, Ku70/80 recognizes DNA broken ends and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form DNA-dependent protein kinase holoenzyme (DNA-PK) in the process of non-homologous end joining (NHEJ). We present a 2.8-Å-resolution cryo-EM structure of DNA-PKcs, allowing precise amino acid sequence registration in regions uninterpreted in previous 4.3-Å X-ray maps. We also report a cryo-EM structure of DNA-PK at 3.5-Å resolution and reveal a dimer mediated by the Ku80 C terminus. Central to dimer formation is a domain swap of the conserved C-terminal helix of Ku80. Our results suggest a new mechanism for NHEJ utilizing a DNA-PK dimer to bring broken DNA ends together. Furthermore, drug inhibition of NHEJ in combination with chemo- and radiotherapy has proved successful, making these models central to structure-based drug targeting efforts.