Summary information and primary citation
- PDB-id
- 6zj2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (3.38 Å)
- Summary
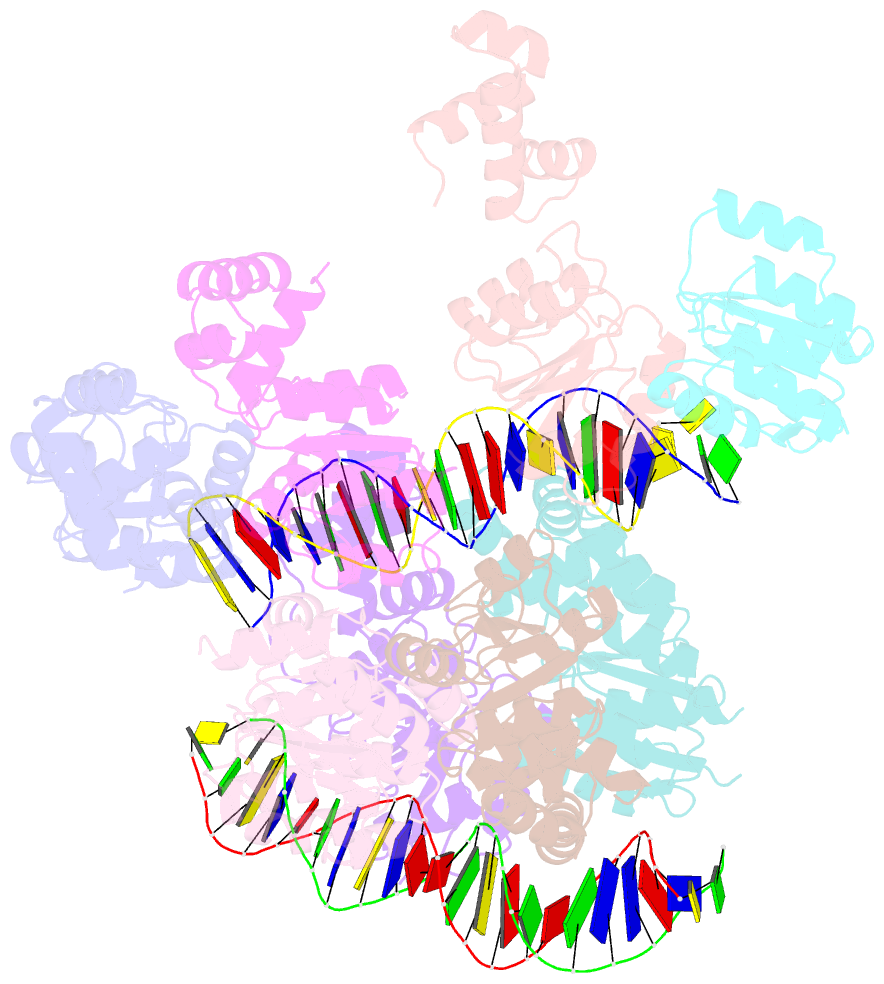
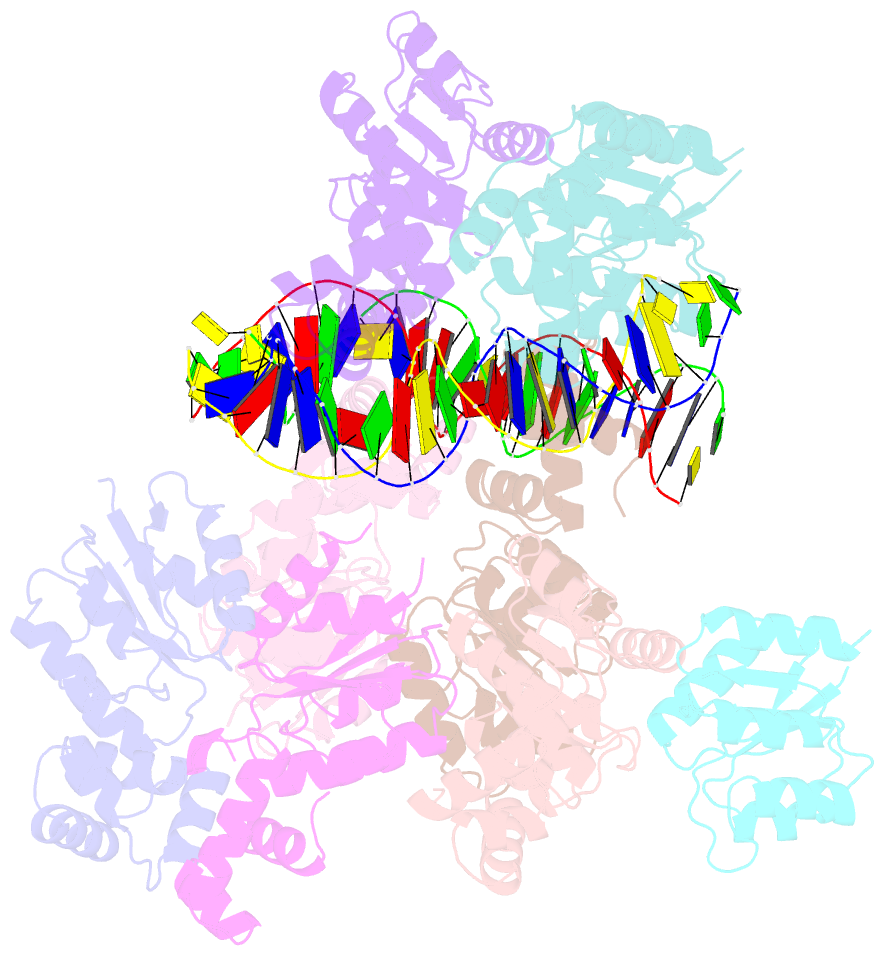
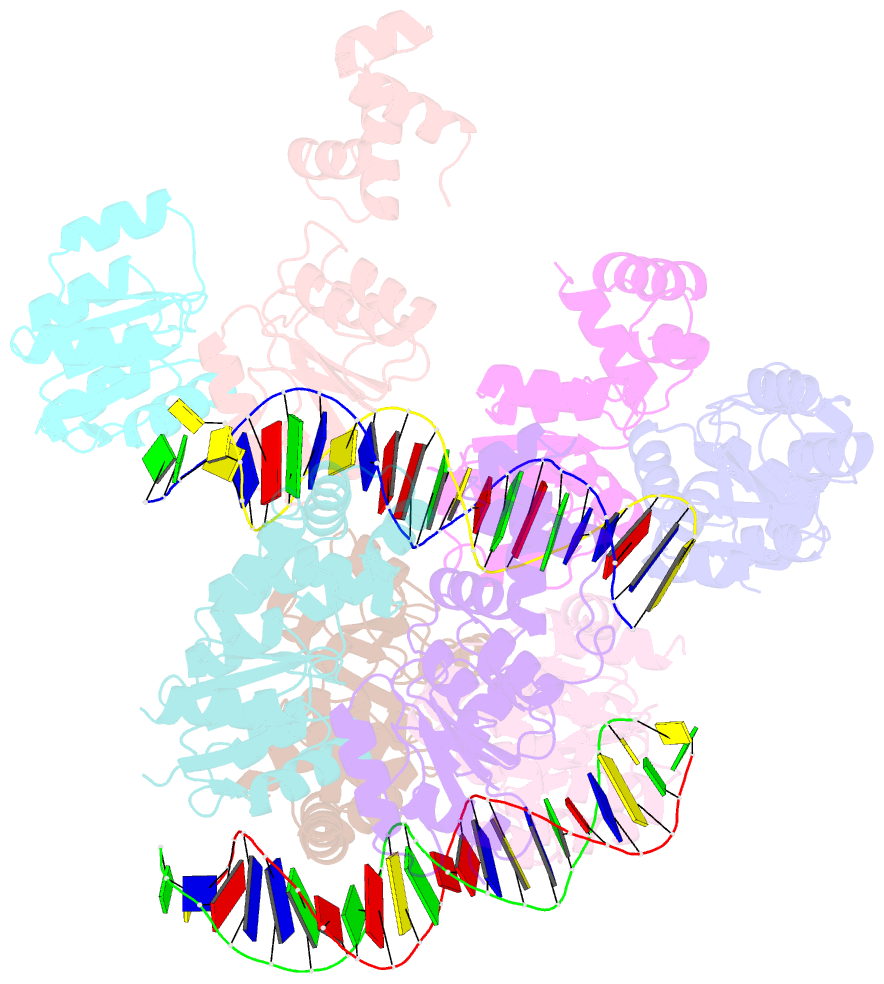
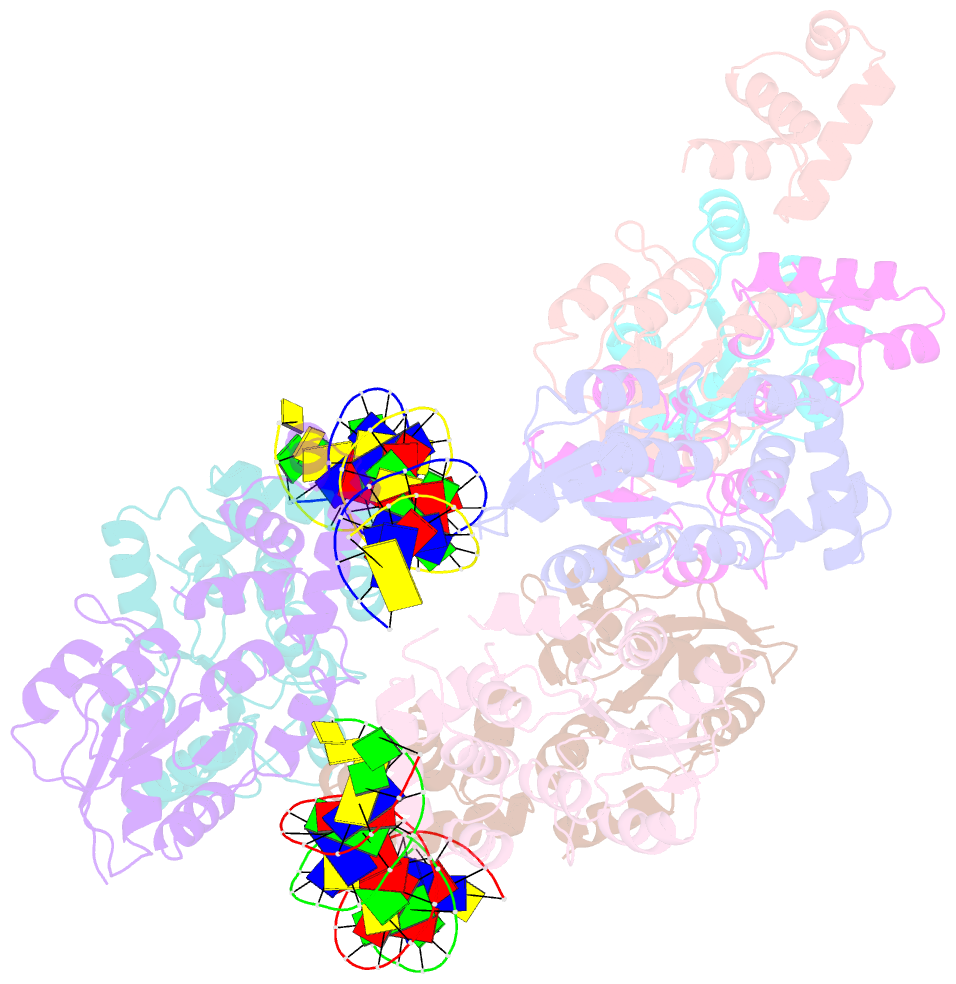
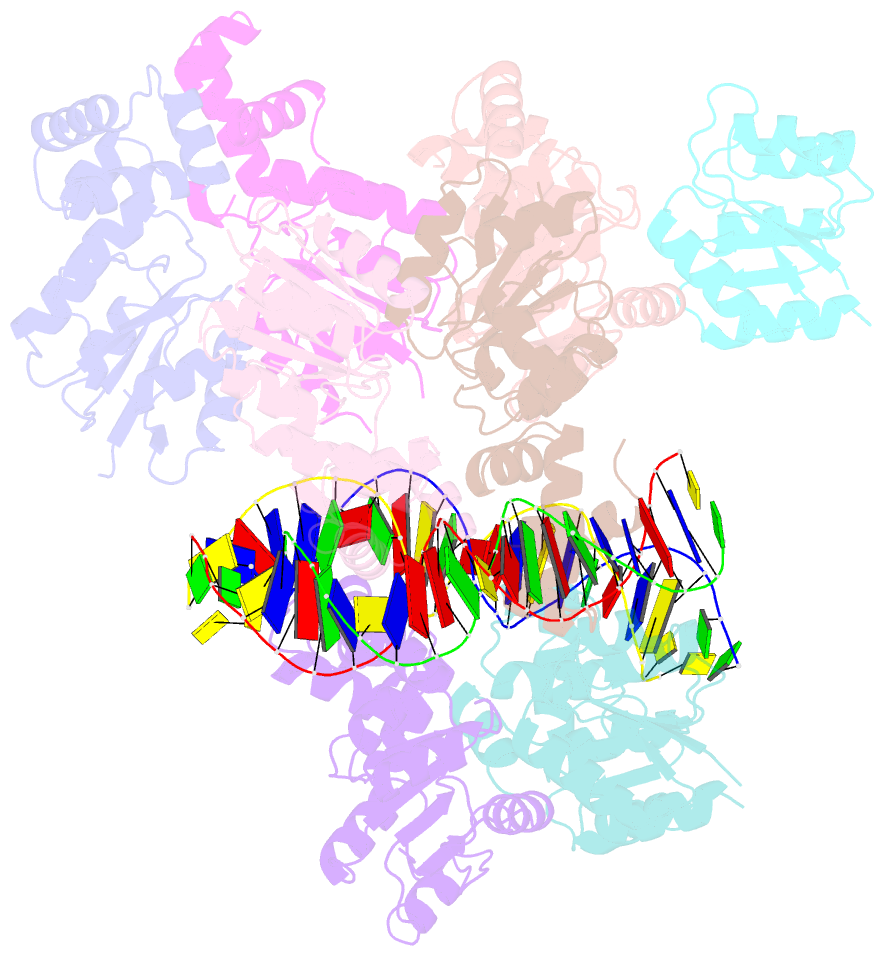
- Structure of rcsb from salmonella enterica serovar typhimurium bound to promoter rpra in the presence of phosphomimetic bef3-
- Reference
- Huesa J, Giner-Lamia J, Pucciarelli MG, Paredes-Martinez F, Portillo FG, Marina A, Casino P (2021): "Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon." Nucleic Acids Res., 49, 2357-2374. doi: 10.1093/nar/gkab060.
- Abstract
- RcsB is a transcriptional regulator that controls expression of numerous genes in enteric bacteria. RcsB accomplishes this role alone or in combination with auxiliary transcriptional factors independently or dependently of phosphorylation. To understand the mechanisms by which RcsB regulates such large number of genes, we performed structural studies as well as in vitro and in vivo functional studies with different RcsB variants. Our structural data reveal that RcsB binds promoters of target genes such as rprA and flhDC in a dimeric active conformation. In this state, the RcsB homodimer docks the DNA-binding domains into the major groove of the DNA, facilitating an initial weak read-out of the target sequence. Interestingly, comparative structural analyses also show that DNA binding may stabilize an active conformation in unphosphorylated RcsB. Furthermore, RNAseq performed in strains expressing wild-type or several RcsB variants provided new insights into the contribution of phosphorylation to gene regulation and assign a potential role of RcsB in controlling iron metabolism. Finally, we delimited the RcsB box for homodimeric active binding to DNA as the sequence TN(G/A)GAN4TC(T/C)NA. This RcsB box was found in promoter, intergenic and intragenic regions, facilitating both increased or decreased gene transcription.