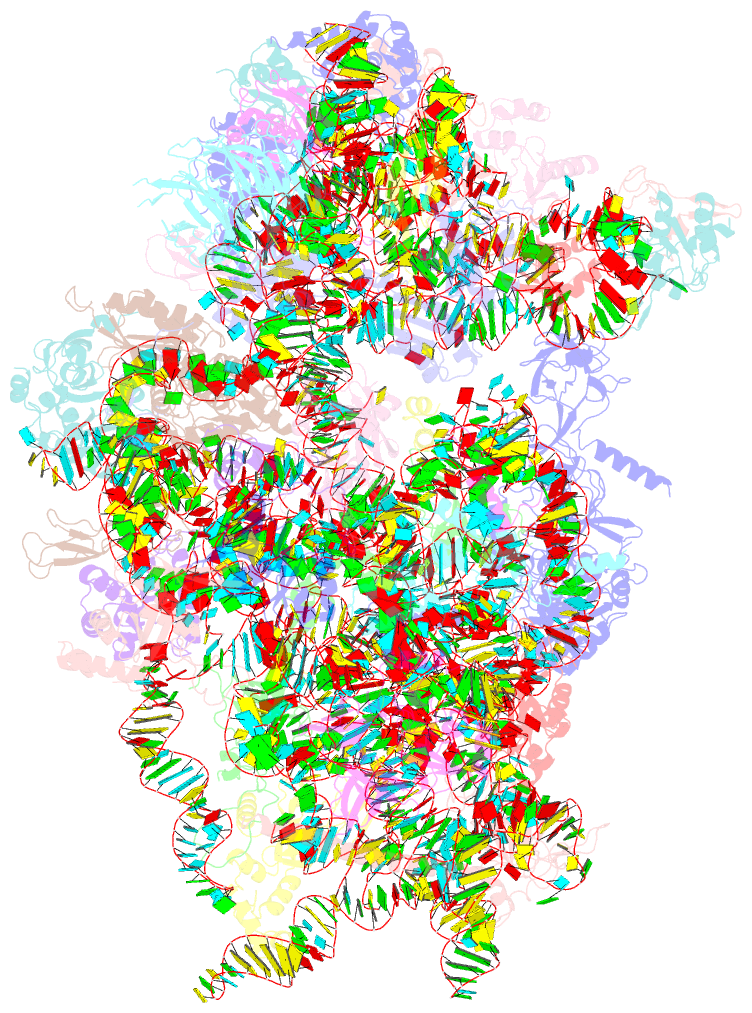
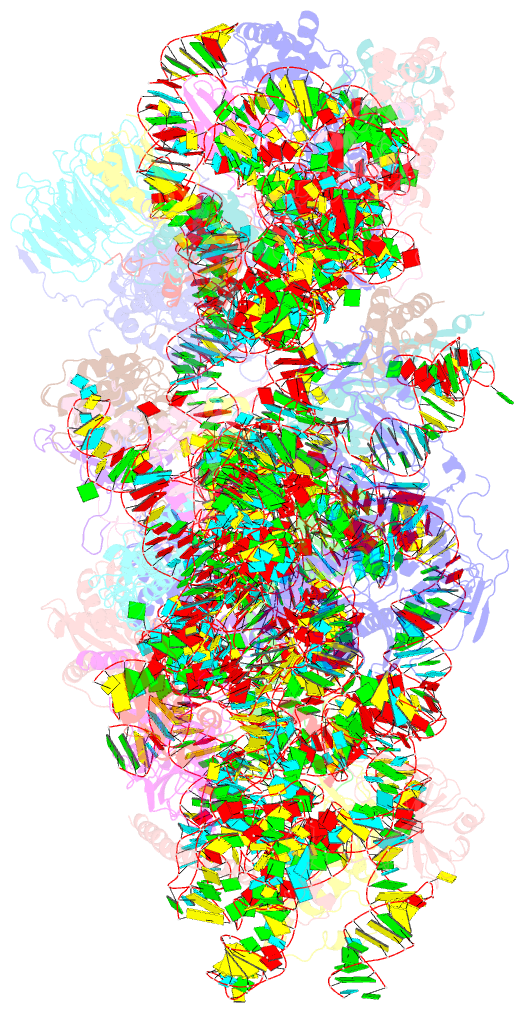
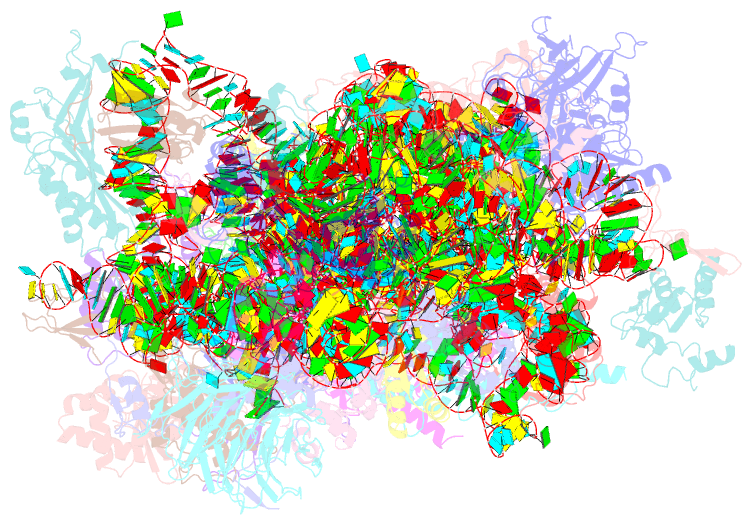
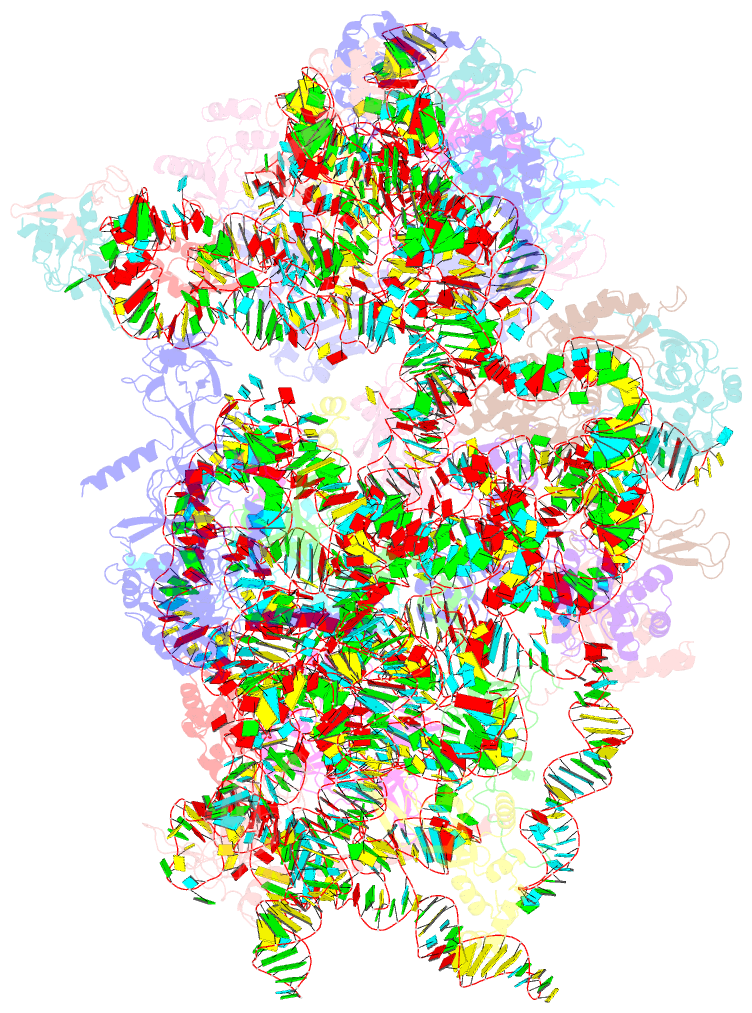
Summary information and primary citation
- PDB-id
- 6zmt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.0 Å)
- Summary
- Sars-cov-2 nsp1 bound to a pre-40s-like ribosome complex
- Reference
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Sturzel CM, Frohlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R (2020): "Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2." Science, 369, 1249-1255. doi: 10.1126/science.abc8665.
- Abstract
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the current coronavirus disease 2019 (COVID-19) pandemic. A major virulence factor of SARS-CoVs is the nonstructural protein 1 (Nsp1), which suppresses host gene expression by ribosome association. Here, we show that Nsp1 from SARS-CoV-2 binds to the 40S ribosomal subunit, resulting in shutdown of messenger RNA (mRNA) translation both in vitro and in cells. Structural analysis by cryo-electron microscopy of in vitro-reconstituted Nsp1-40S and various native Nsp1-40S and -80S complexes revealed that the Nsp1 C terminus binds to and obstructs the mRNA entry tunnel. Thereby, Nsp1 effectively blocks retinoic acid-inducible gene I-dependent innate immune responses that would otherwise facilitate clearance of the infection. Thus, the structural characterization of the inhibitory mechanism of Nsp1 may aid structure-based drug design against SARS-CoV-2.