Summary information and primary citation
- PDB-id
- 7a08; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- cryo-EM (3.11 Å)
- Summary
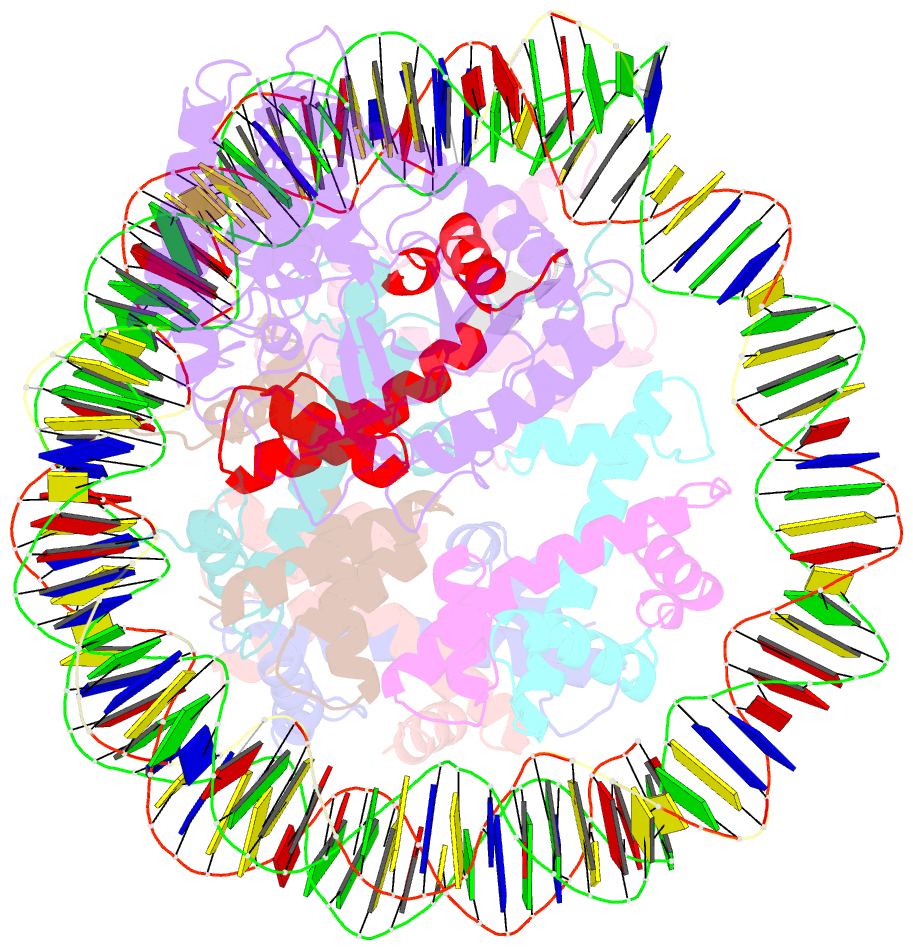
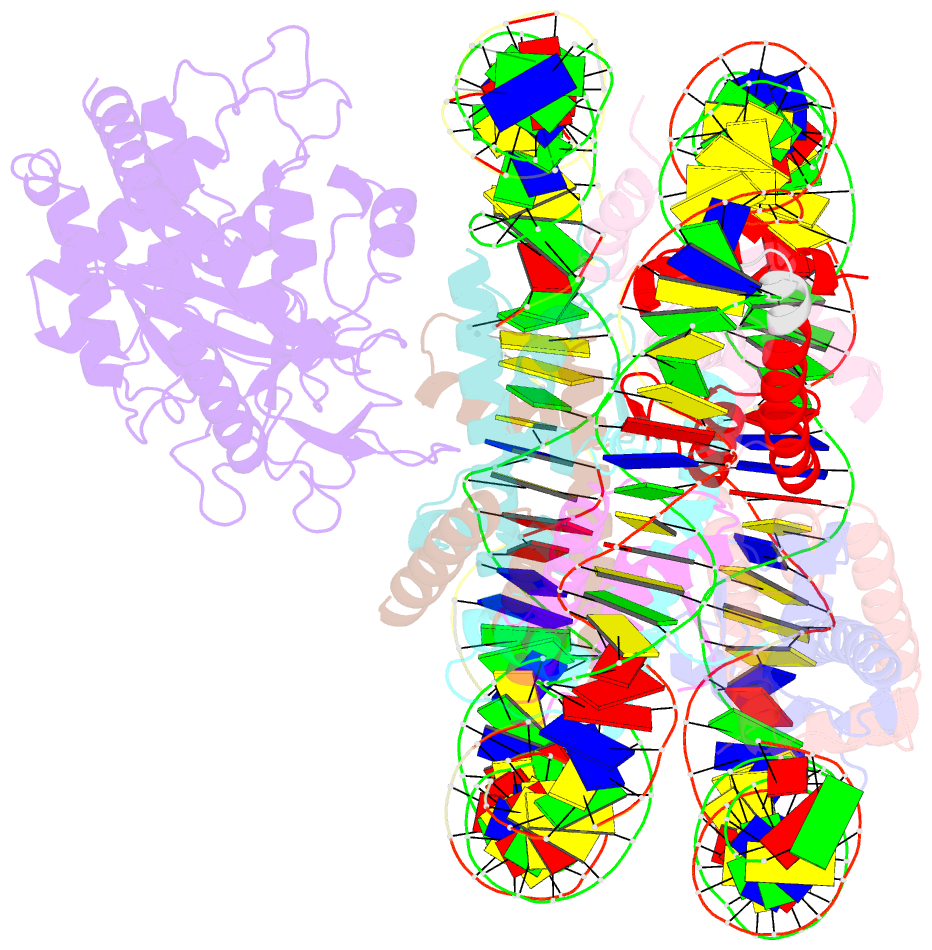
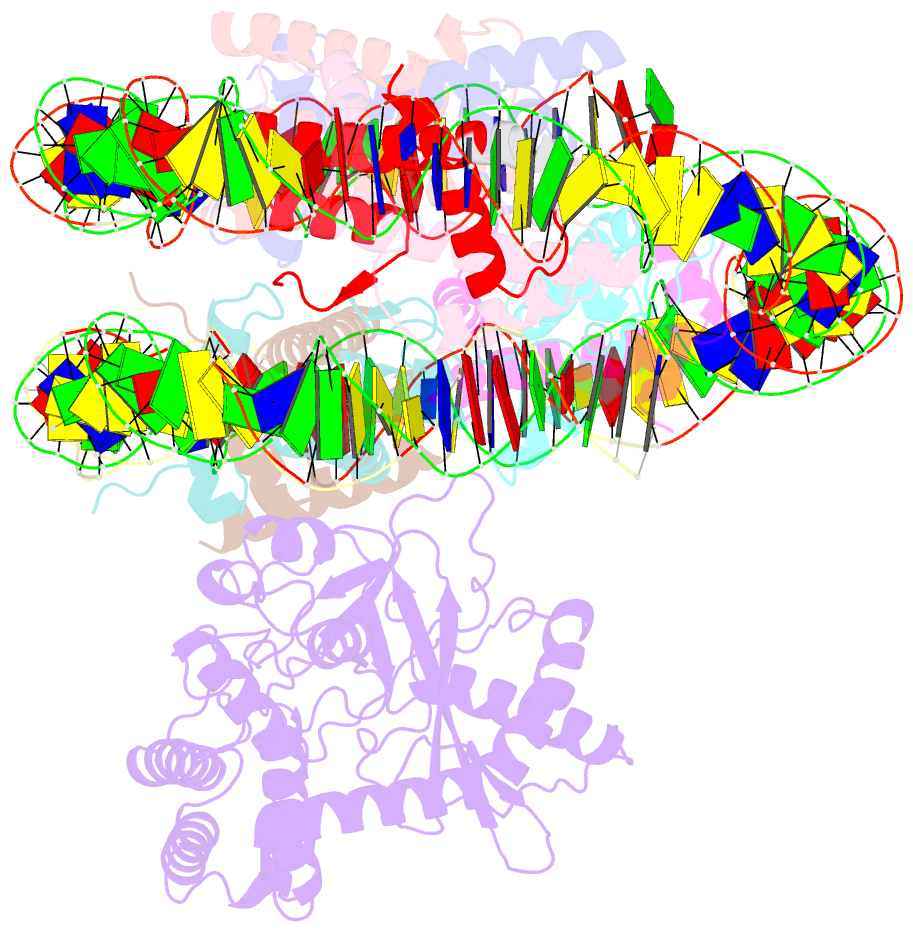
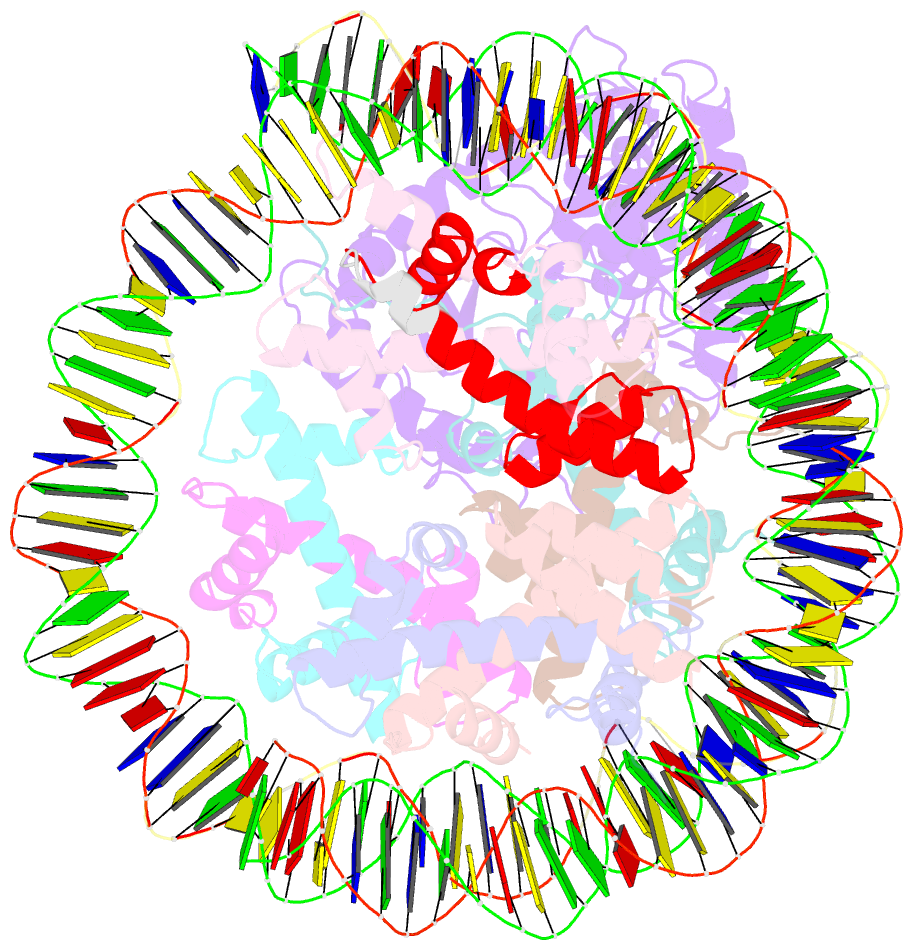
- Cryoem structure of cgas nucleosome complex
- Reference
- Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, Hornung V, Hopfner KP (2020): "Structural basis for sequestration and autoinhibition of cGAS by chromatin." Nature, 587, 678-682. doi: 10.1038/s41586-020-2748-0.
- Abstract
- Cyclic GMP-AMP synthase (cGAS) is an innate immune sensor for cytosolic microbial DNA1. After binding DNA, cGAS synthesizes the messenger 2'3'-cyclic GMP-AMP (cGAMP)2-4, which triggers cell-autonomous defence and the production of type I interferons and pro-inflammatory cytokines via the activation of STING5. In addition to responding to cytosolic microbial DNA, cGAS also recognizes mislocalized cytosolic self-DNA and has been implicated in autoimmunity and sterile inflammation6,7. Specificity towards pathogen- or damage-associated DNA was thought to be caused by cytosolic confinement. However, recent findings place cGAS robustly in the nucleus8-10, where tight tethering of chromatin is important to prevent autoreactivity to self-DNA8. Here we show how cGAS is sequestered and inhibited by chromatin. We provide a cryo-electron microscopy structure of the cGAS catalytic domain bound to a nucleosome, which shows that cGAS does not interact with the nucleosomal DNA, but instead interacts with histone 2A-histone 2B, and is tightly anchored to the 'acidic patch'. The interaction buries the cGAS DNA-binding site B, and blocks the formation of active cGAS dimers. The acidic patch robustly outcompetes agonistic DNA for binding to cGAS, which suggests that nucleosome sequestration can efficiently inhibit cGAS, even when accessible DNA is nearby, such as in actively transcribed genomic regions. Our results show how nuclear cGAS is sequestered by chromatin and provides a mechanism for preventing autoreactivity to nuclear self-DNA.