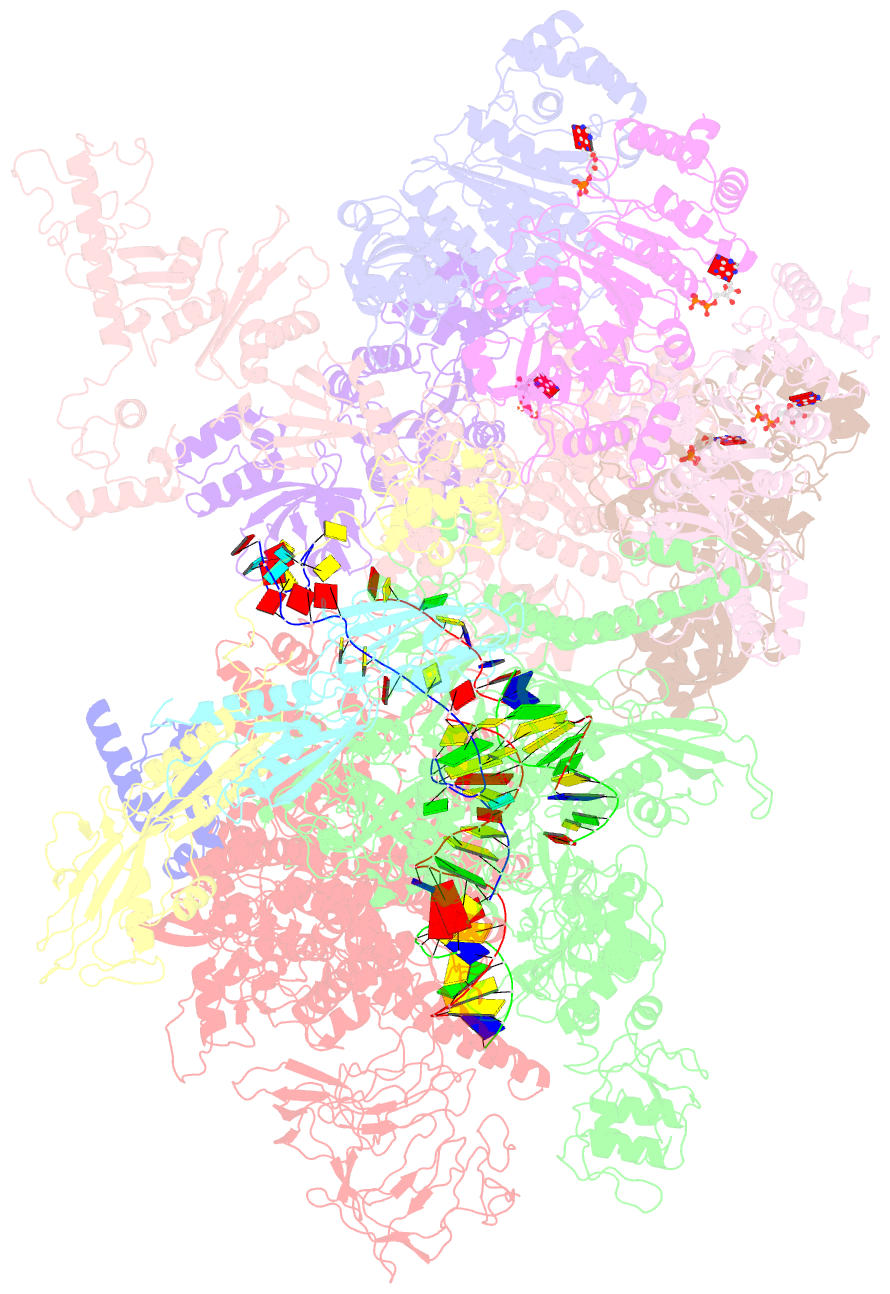
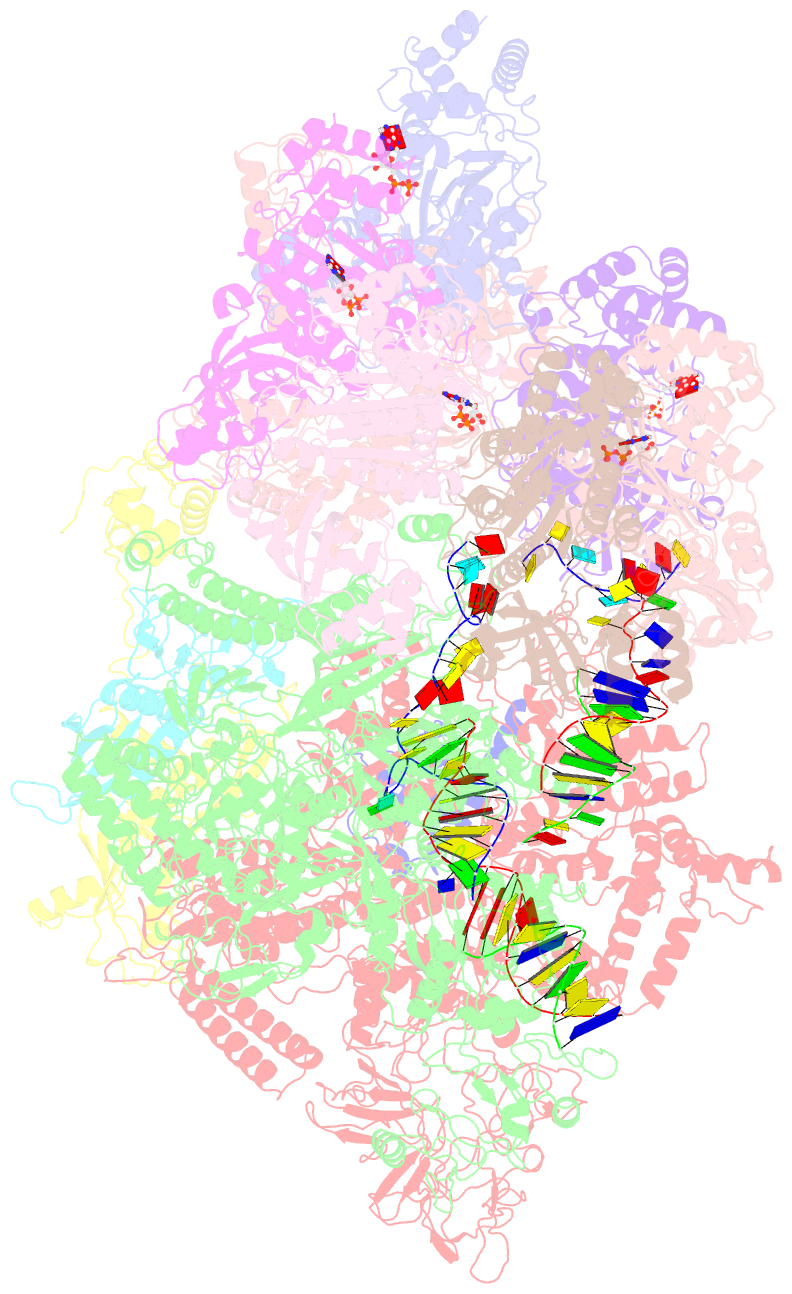
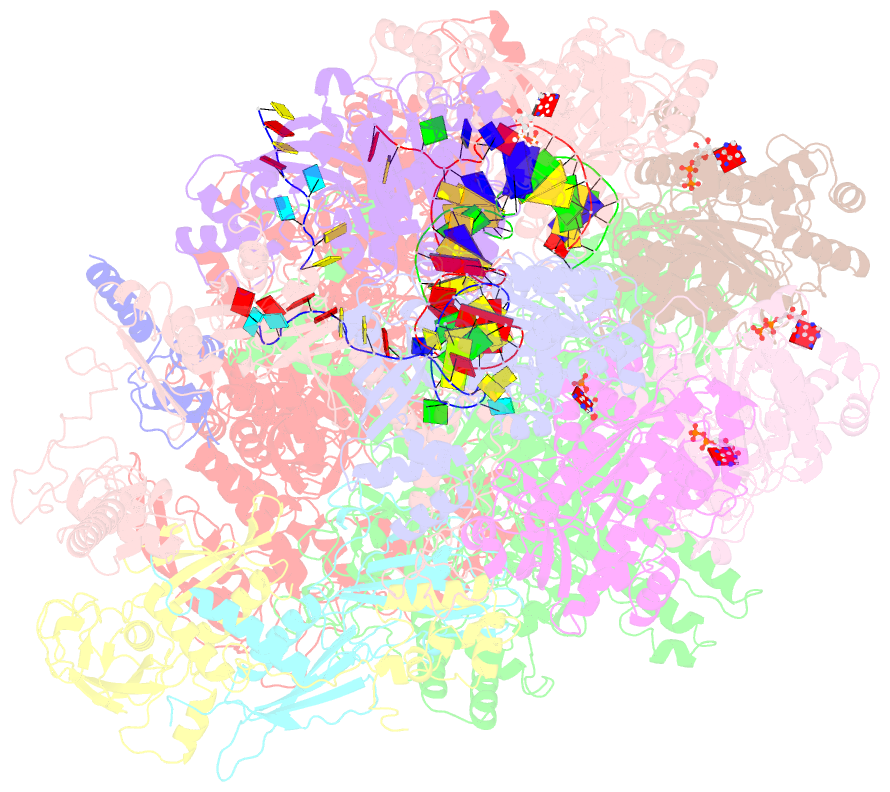
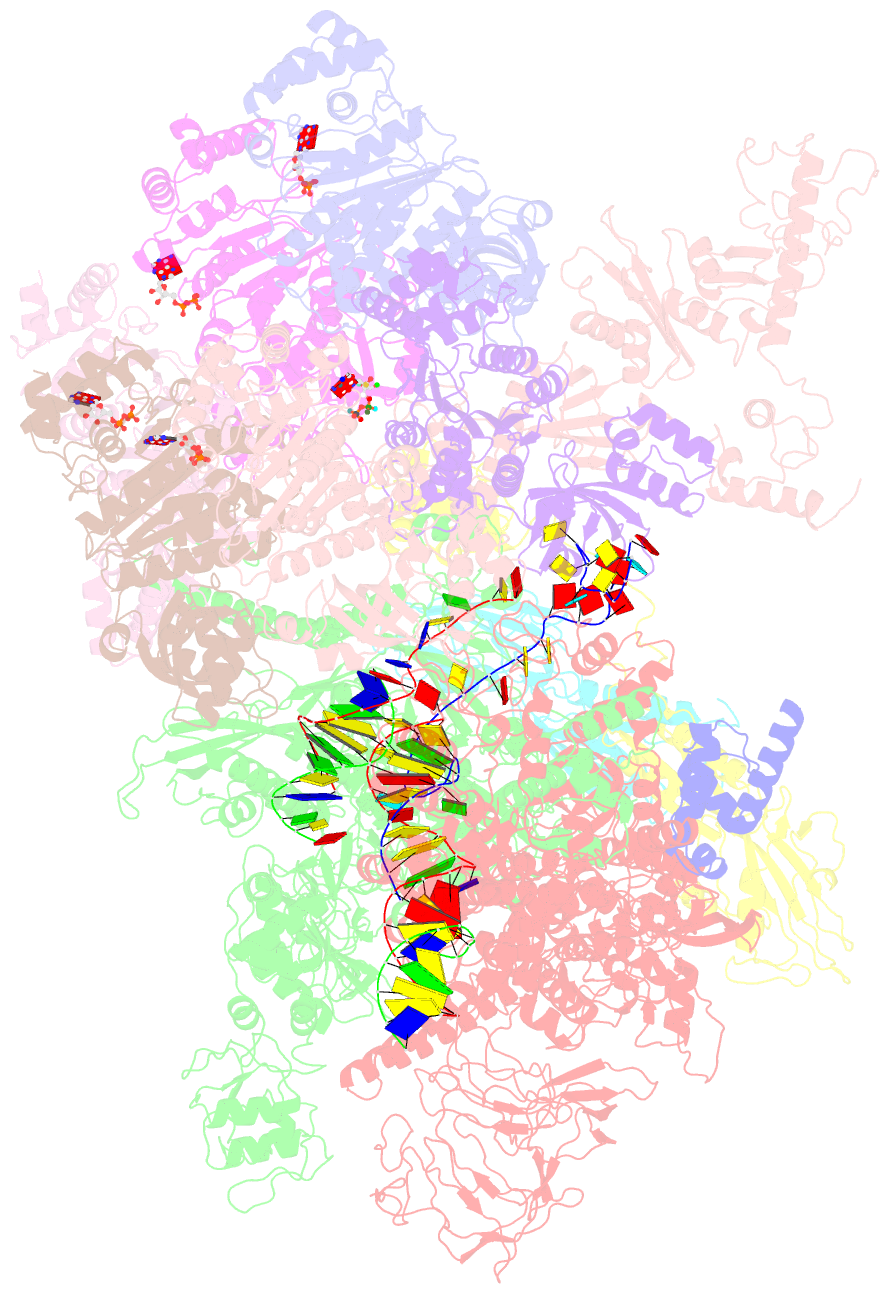
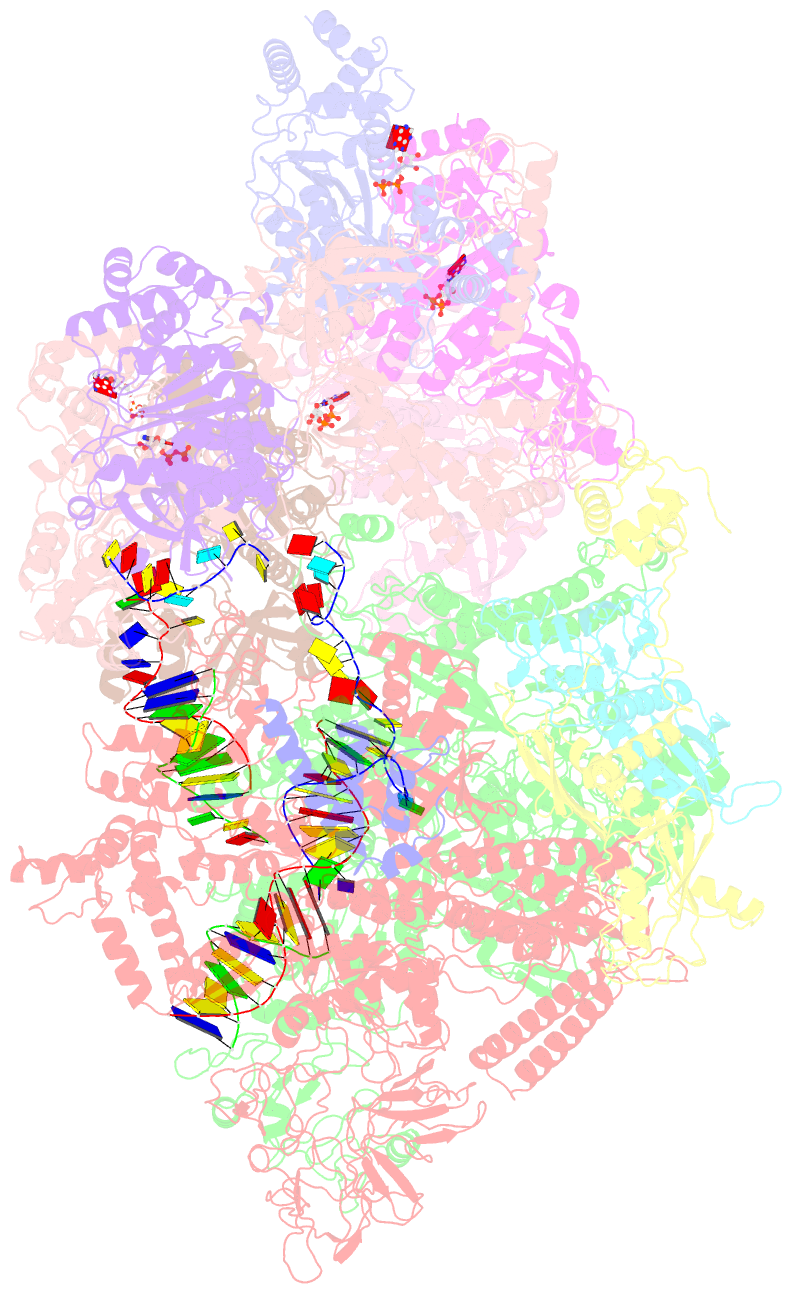
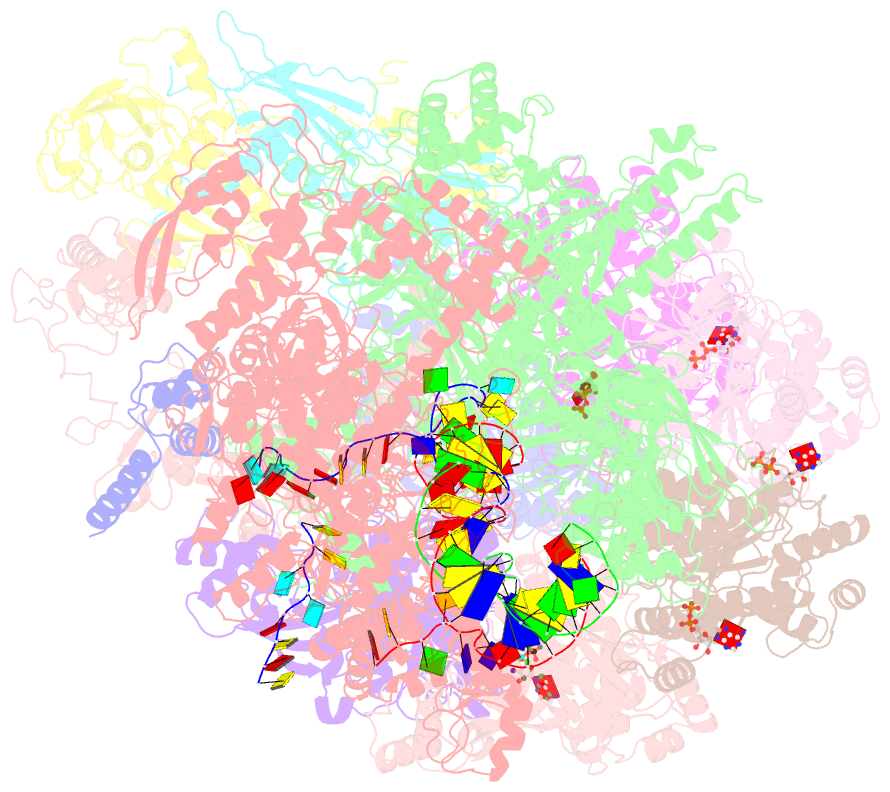
Summary information and primary citation
- PDB-id
- 7ade; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.2 Å)
- Summary
- Transcription termination complex iva
- Reference
- Said N, Hilal T, Sunday ND, Khatri A, Burger J, Mielke T, Belogurov GA, Loll B, Sen R, Artsimovitch I, Wahl MC (2021): "Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho." Science, 371. doi: 10.1126/science.abd1673.
- Abstract
- Factor-dependent transcription termination mechanisms are poorly understood. We determined a series of cryo-electron microscopy structures portraying the hexameric adenosine triphosphatase (ATPase) ρ on a pathway to terminating NusA/NusG-modified elongation complexes. An open ρ ring contacts NusA, NusG, and multiple regions of RNA polymerase, trapping and locally unwinding proximal upstream DNA. NusA wedges into the ρ ring, initially sequestering RNA. Upon deflection of distal upstream DNA over the RNA polymerase zinc-binding domain, NusA rotates underneath one capping ρ subunit, which subsequently captures RNA. After detachment of NusG and clamp opening, RNA polymerase loses its grip on the RNA:DNA hybrid and is inactivated. Our structural and functional analyses suggest that ρ, and other termination factors across life, may use analogous strategies to allosterically trap transcription complexes in a moribund state.