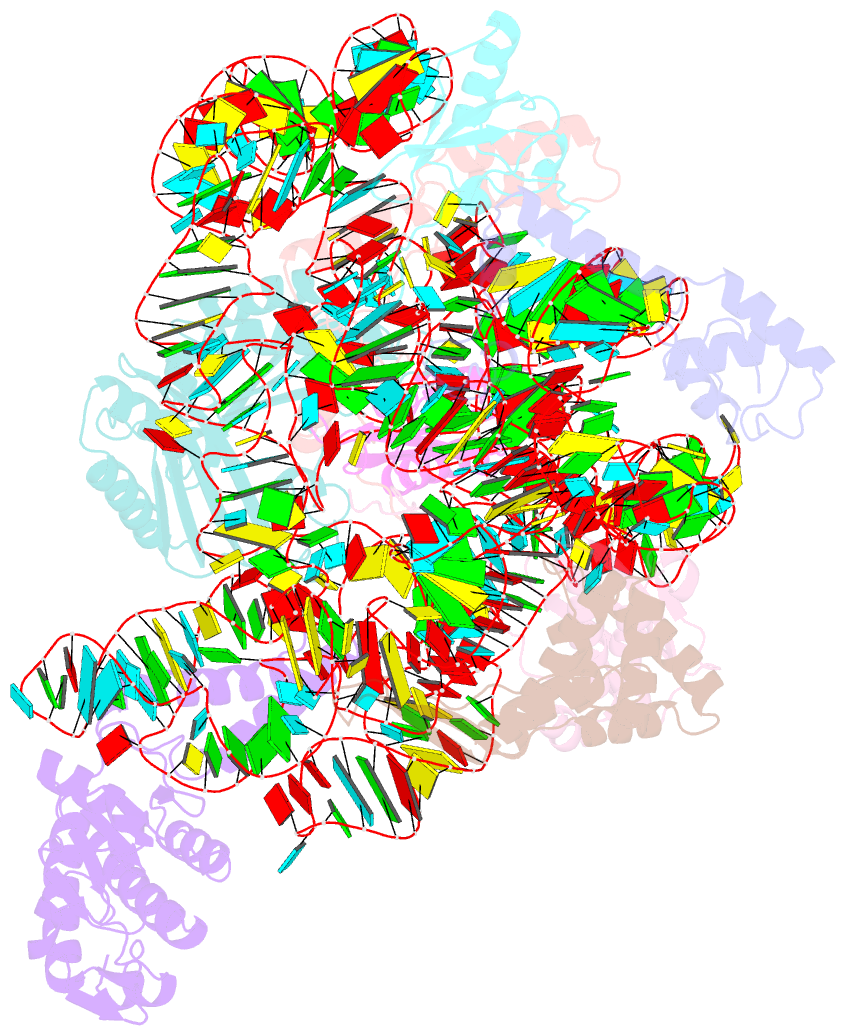
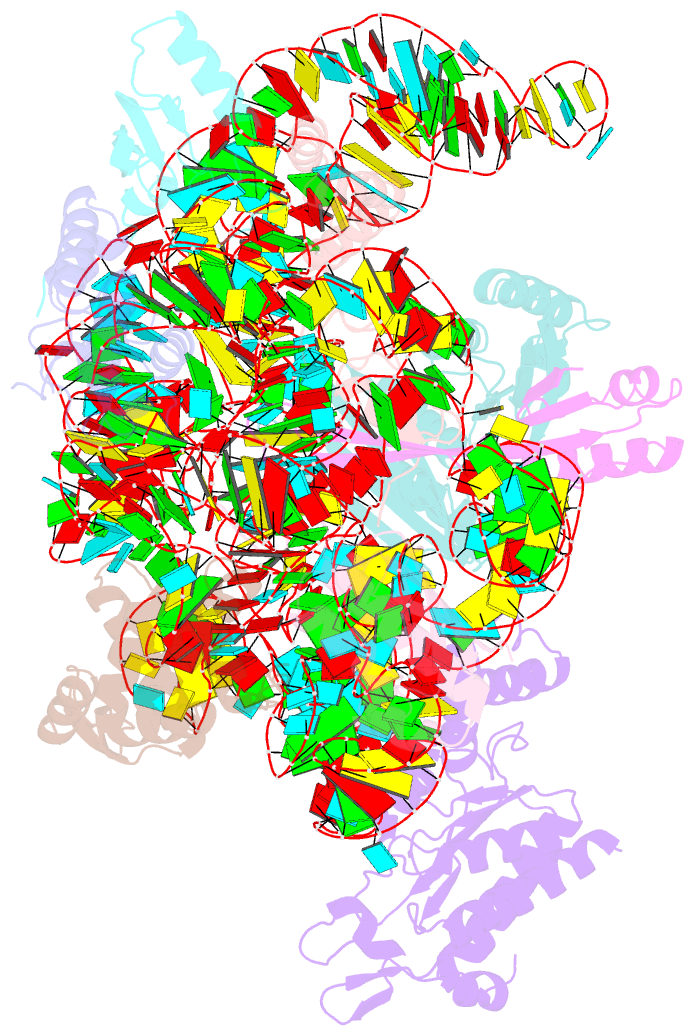
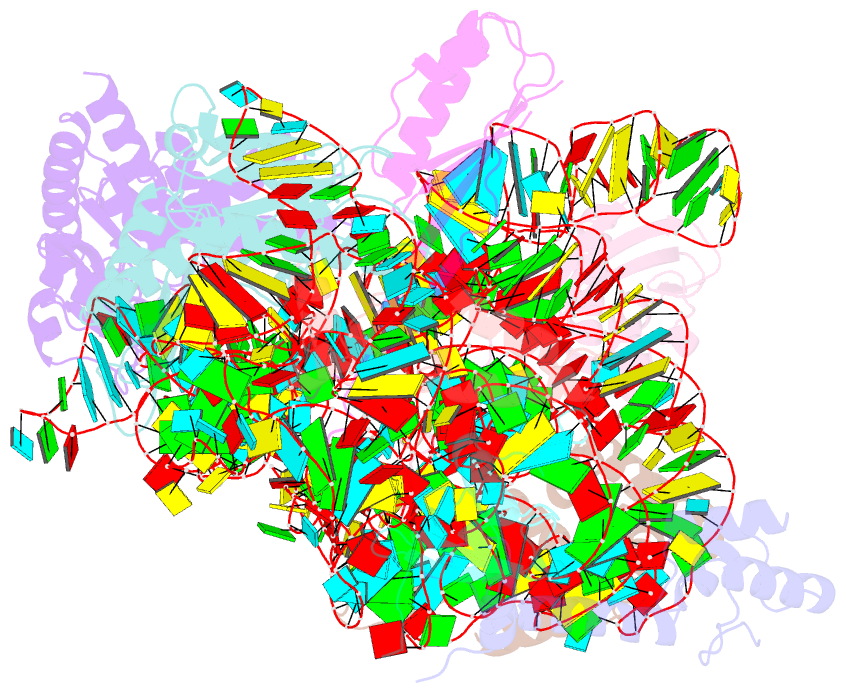
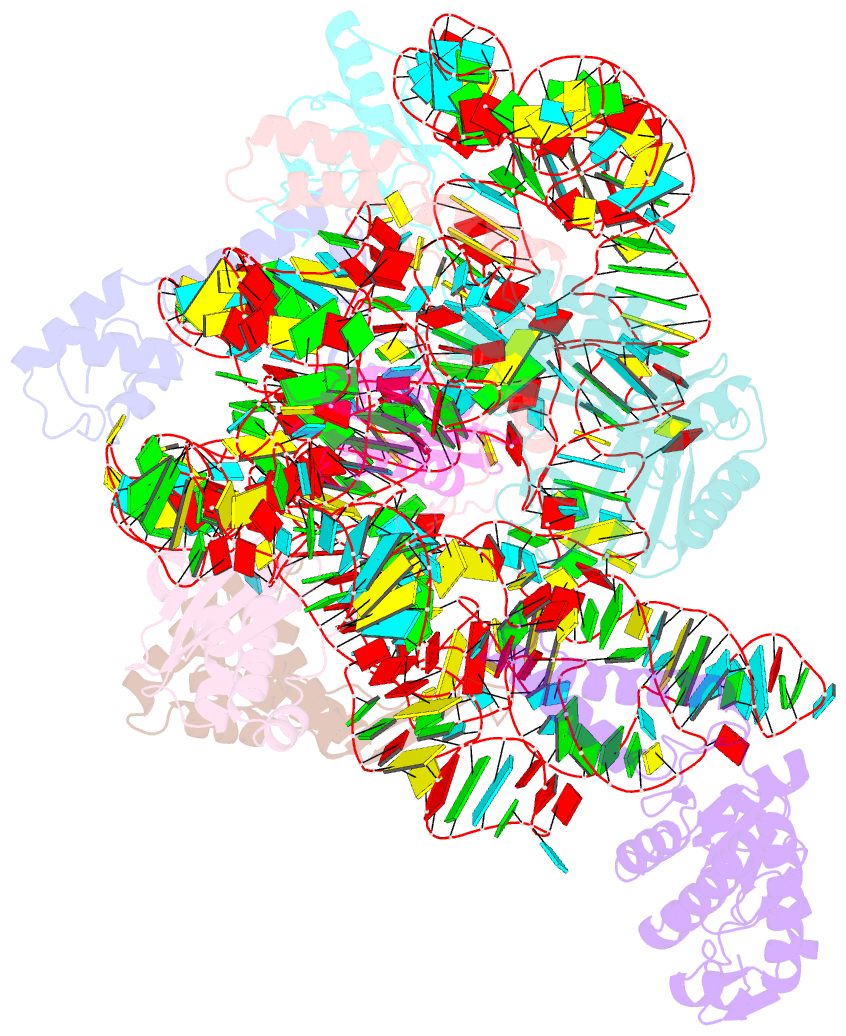
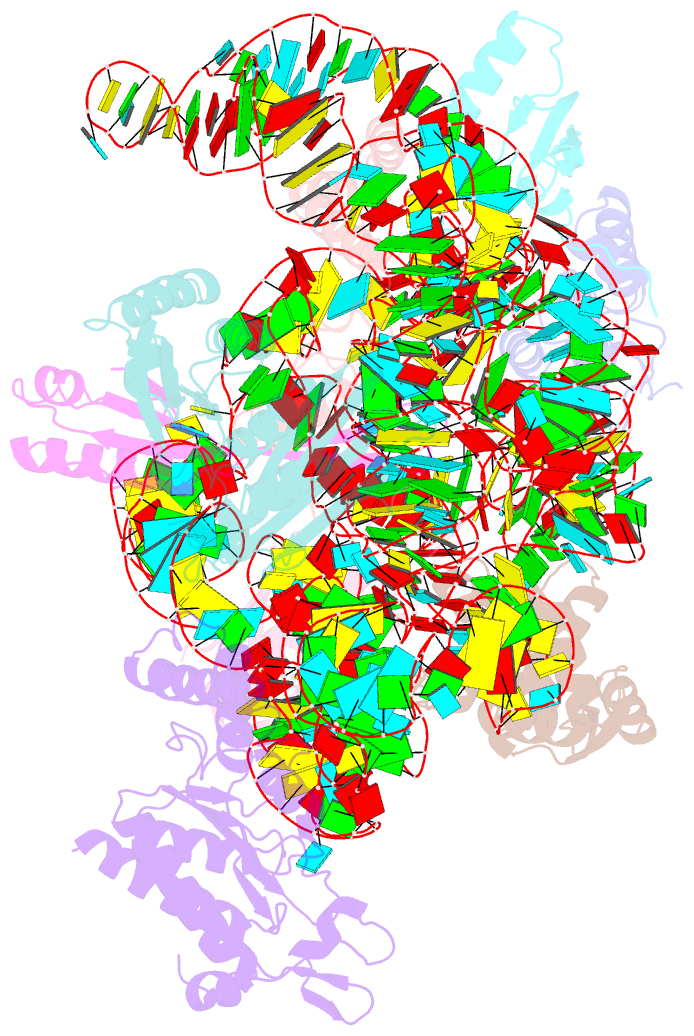
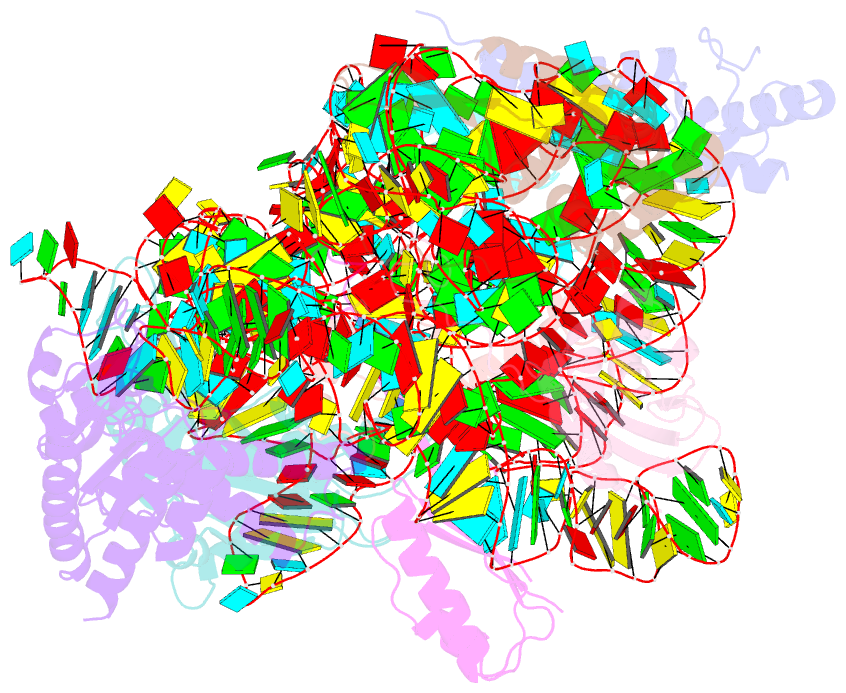
Summary information and primary citation
- PDB-id
- 7afa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.95 Å)
- Summary
- Bacterial 30s ribosomal subunit assembly complex state f (head domain)
- Reference
- Schedlbauer A, Iturrioz I, Ochoa-Lizarralde B, Diercks T, Lopez-Alonso JP, Lavin JL, Kaminishi T, Capuni R, Dhimole N, de Astigarraga E, Gil-Carton D, Fucini P, Connell SR (2021): "A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit." Sci Adv, 7. doi: 10.1126/sciadv.abf7547.
- Abstract
- While a structural description of the molecular mechanisms guiding ribosome assembly in eukaryotic systems is emerging, bacteria use an unrelated core set of assembly factors for which high-resolution structural information is still missing. To address this, we used single-particle cryo-electron microscopy to visualize the effects of bacterial ribosome assembly factors RimP, RbfA, RsmA, and RsgA on the conformational landscape of the 30S ribosomal subunit and obtained eight snapshots representing late steps in the folding of the decoding center. Analysis of these structures identifies a conserved secondary structure switch in the 16S ribosomal RNA central to decoding site maturation and suggests both a sequential order of action and molecular mechanisms for the assembly factors in coordinating and controlling this switch. Structural and mechanistic parallels between bacterial and eukaryotic systems indicate common folding features inherent to all ribosomes.