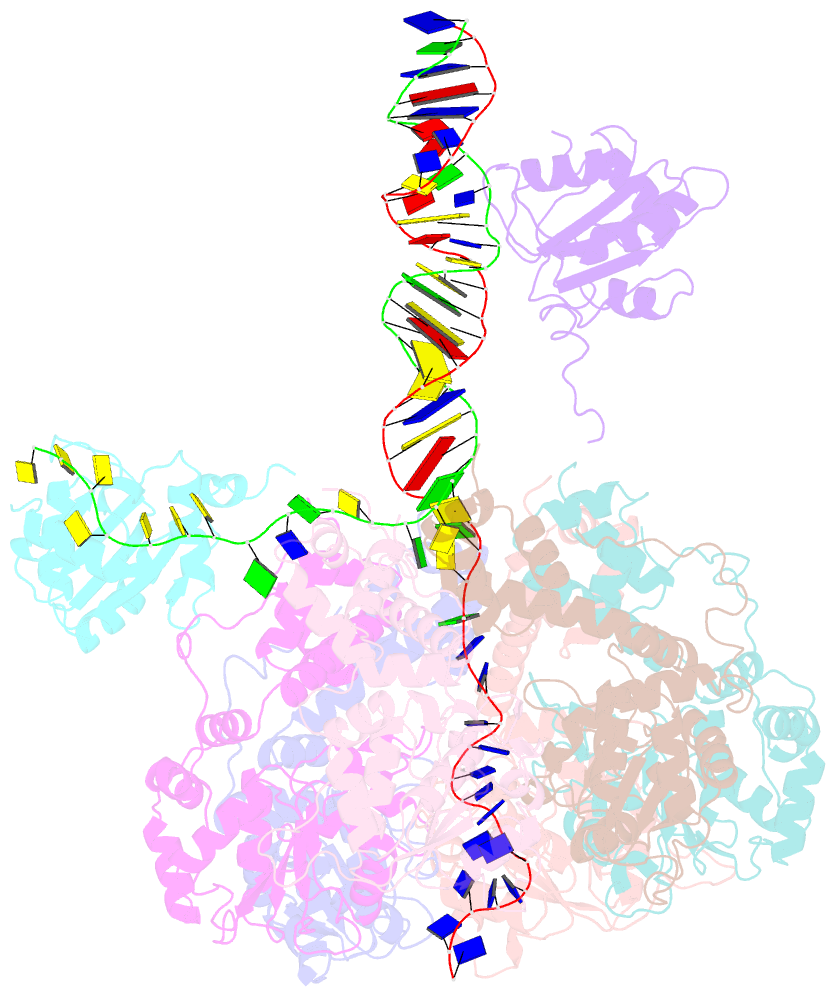
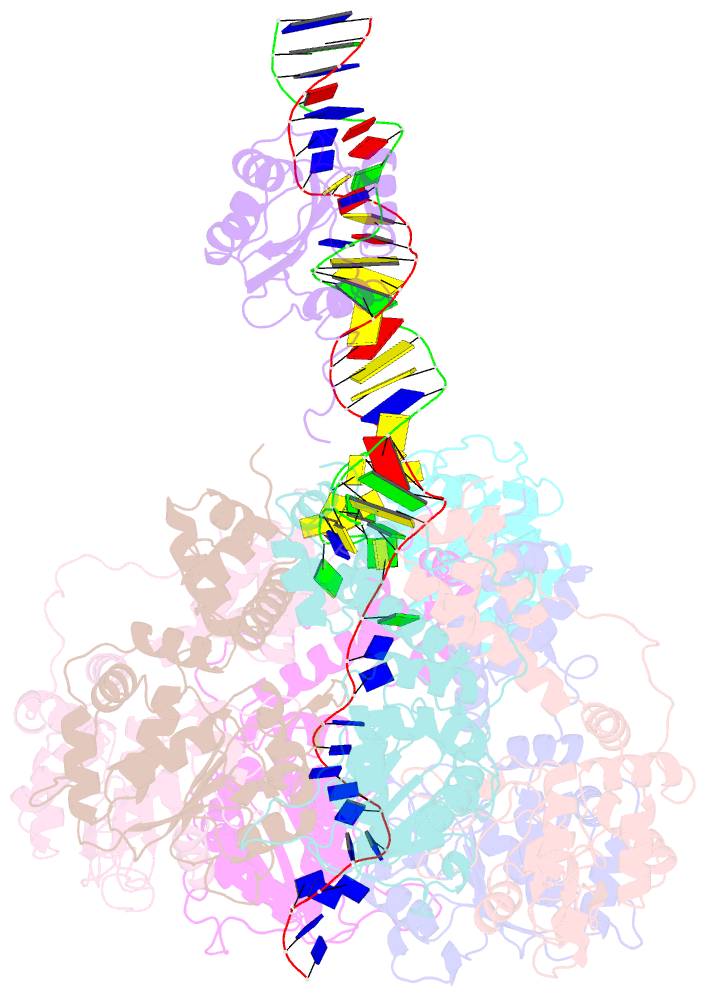
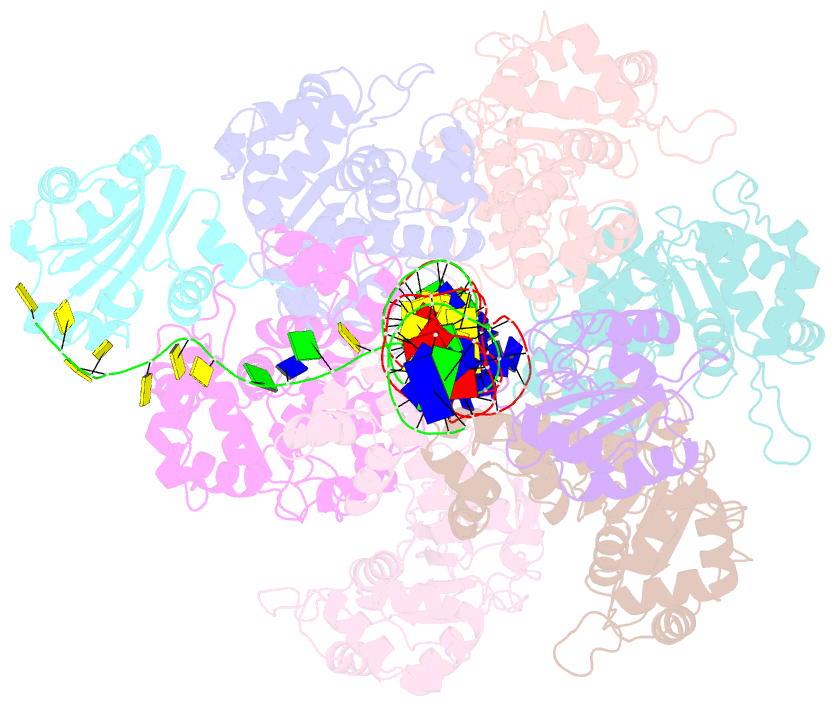
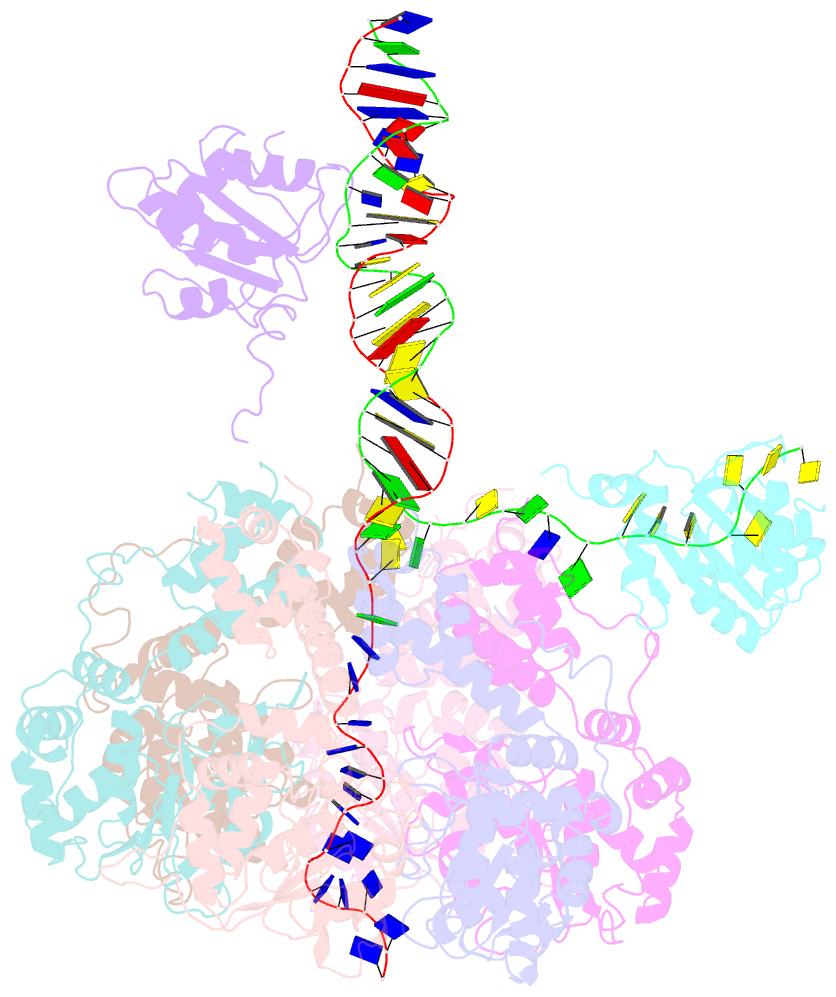
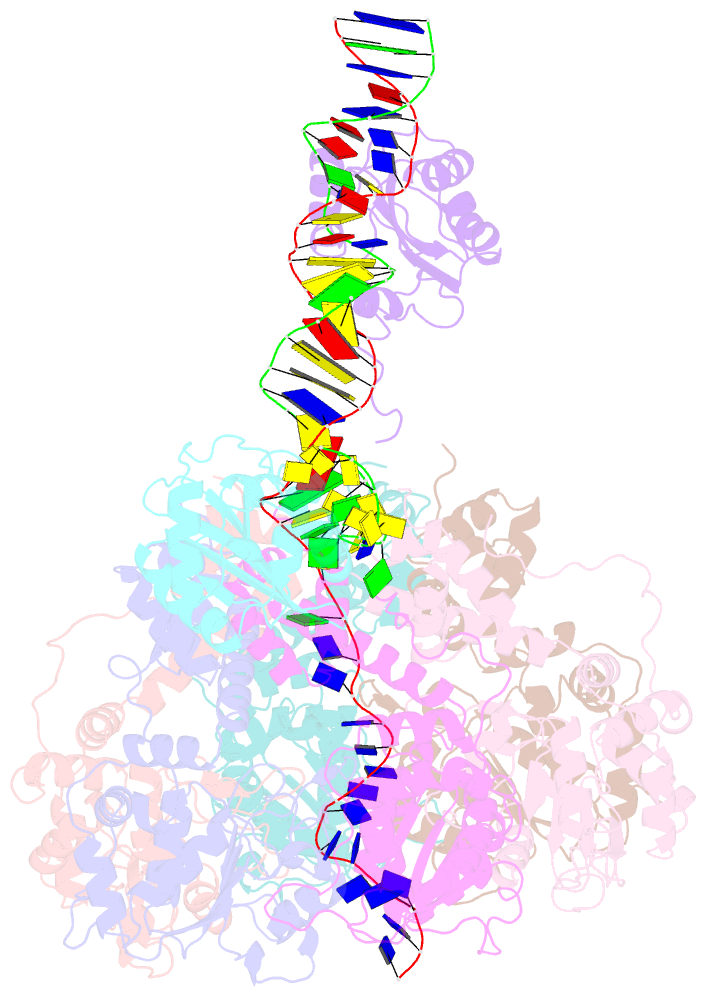
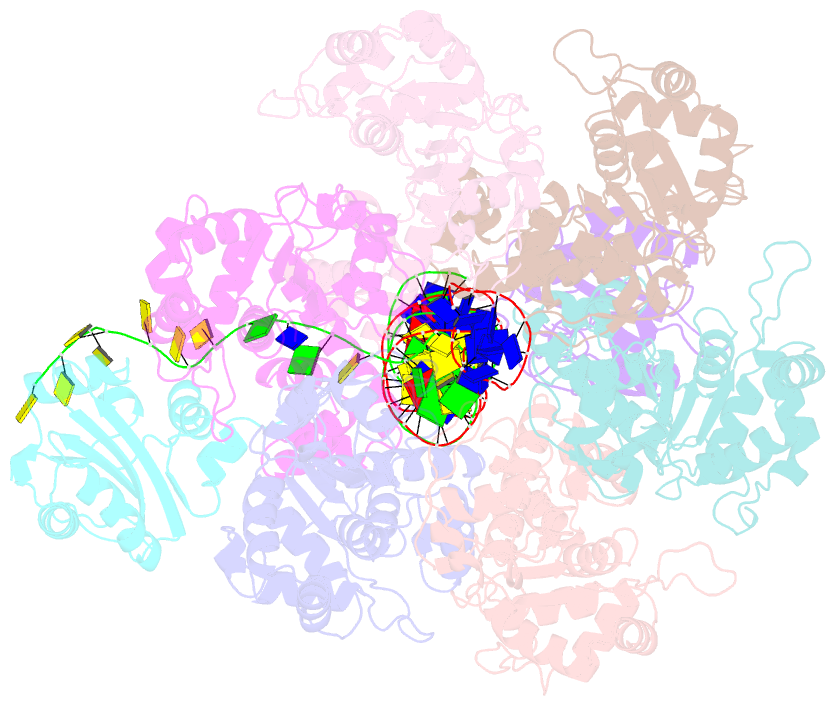
Summary information and primary citation
- PDB-id
- 7apd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.9 Å)
- Summary
- Bovine papillomavirus e1 DNA helicase-replication fork complex
- Reference
- Javed A, Major B, Stead JA, Sanders CM, Orlova EV (2021): "Unwinding of a DNA replication fork by a hexameric viral helicase." Nat Commun, 12, 5535. doi: 10.1038/s41467-021-25843-6.
- Abstract
- Hexameric helicases are motor proteins that unwind double-stranded DNA (dsDNA) during DNA replication but how they are optimised for strand separation is unclear. Here we present the cryo-EM structure of the full-length E1 helicase from papillomavirus, revealing all arms of a bound DNA replication fork and their interactions with the helicase. The replication fork junction is located at the entrance to the helicase collar ring, that sits above the AAA + motor assembly. dsDNA is escorted to and the 5´ single-stranded DNA (ssDNA) away from the unwinding point by the E1 dsDNA origin binding domains. The 3´ ssDNA interacts with six spirally-arranged β-hairpins and their cyclical top-to-bottom movement pulls the ssDNA through the helicase. Pulling of the RF against the collar ring separates the base-pairs, while modelling of the conformational cycle suggest an accompanying movement of the collar ring has an auxiliary role, helping to make efficient use of ATP in duplex unwinding.