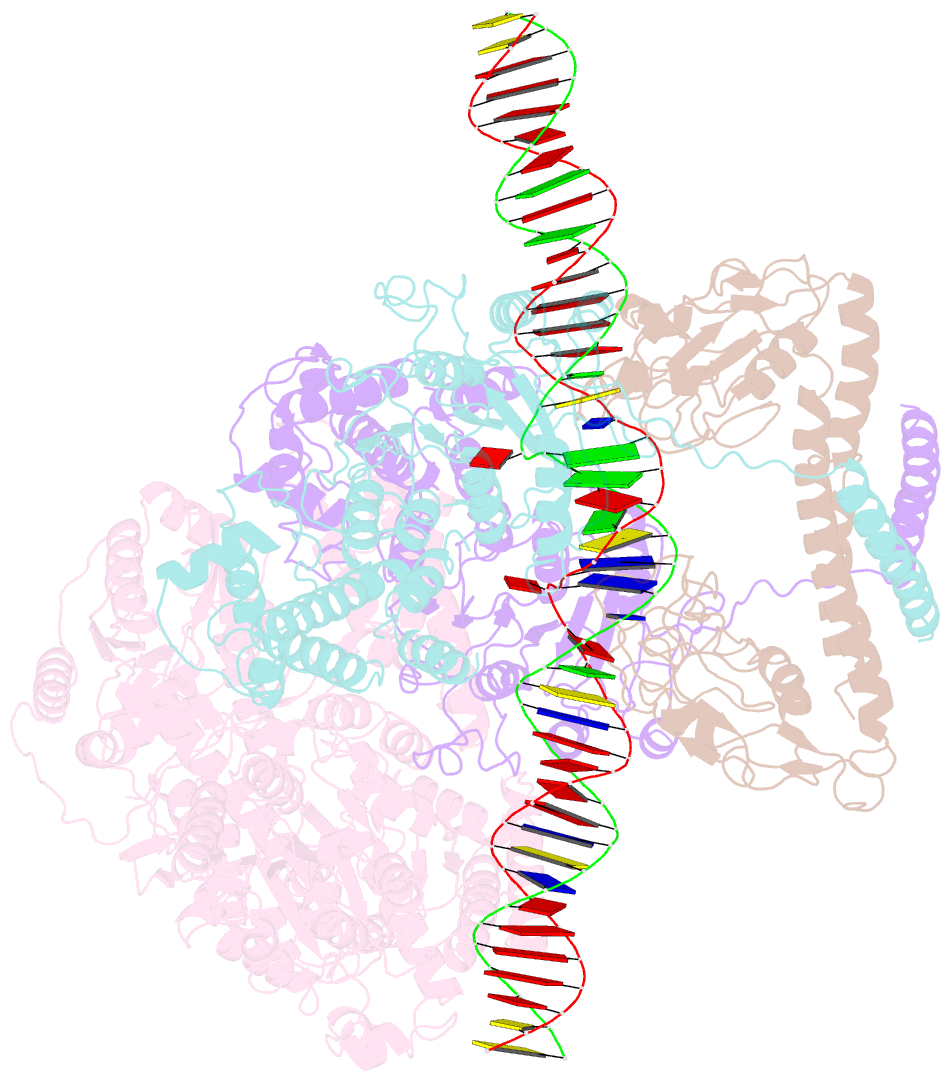
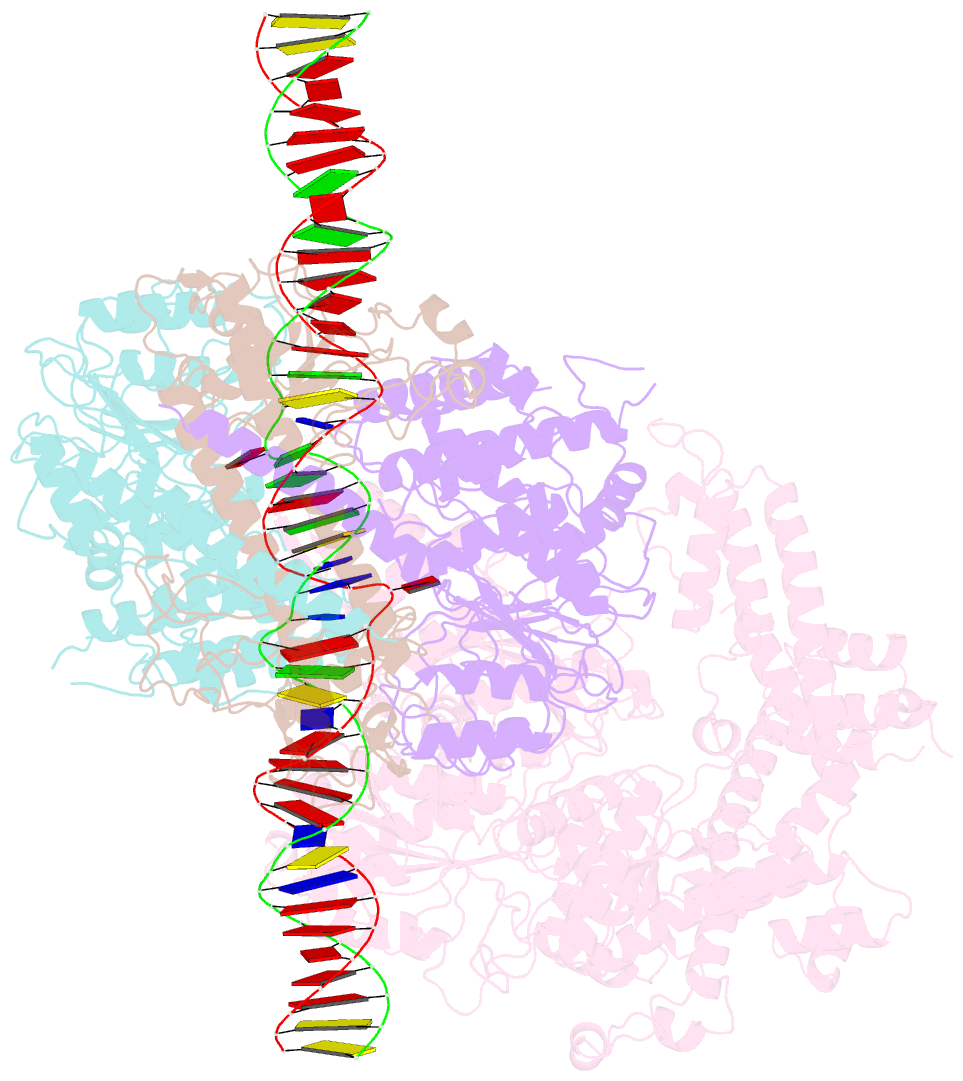
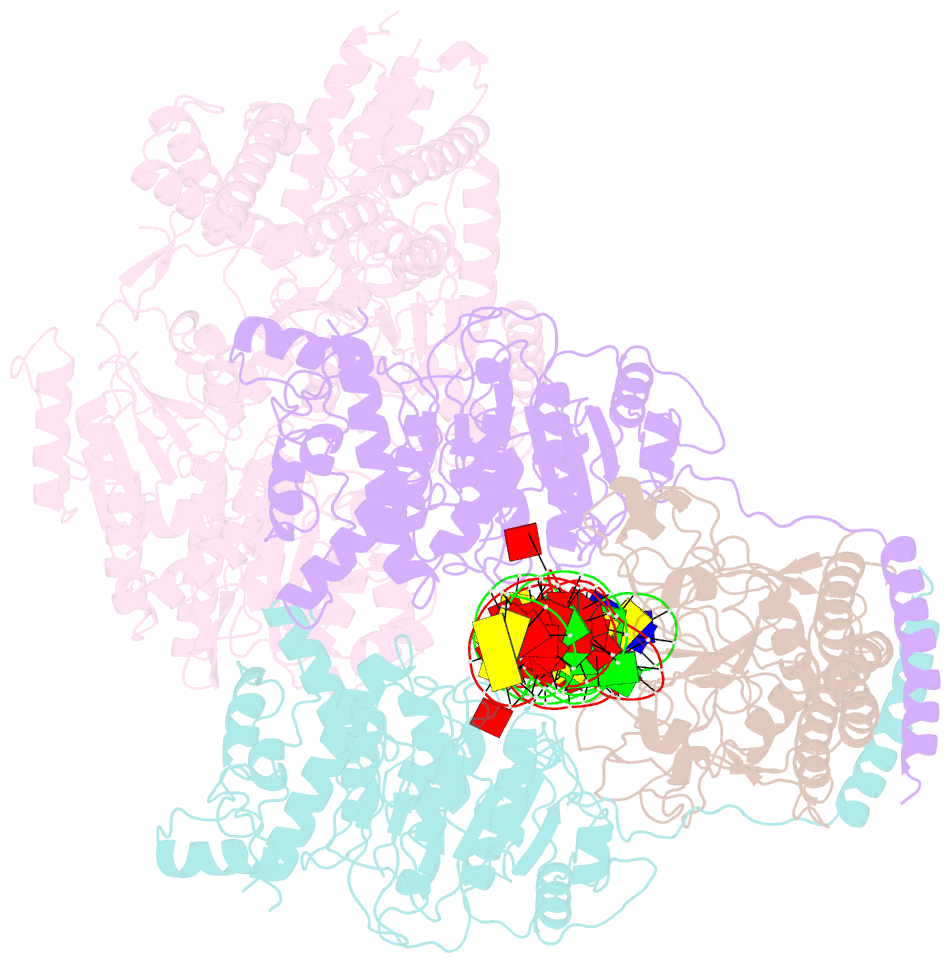
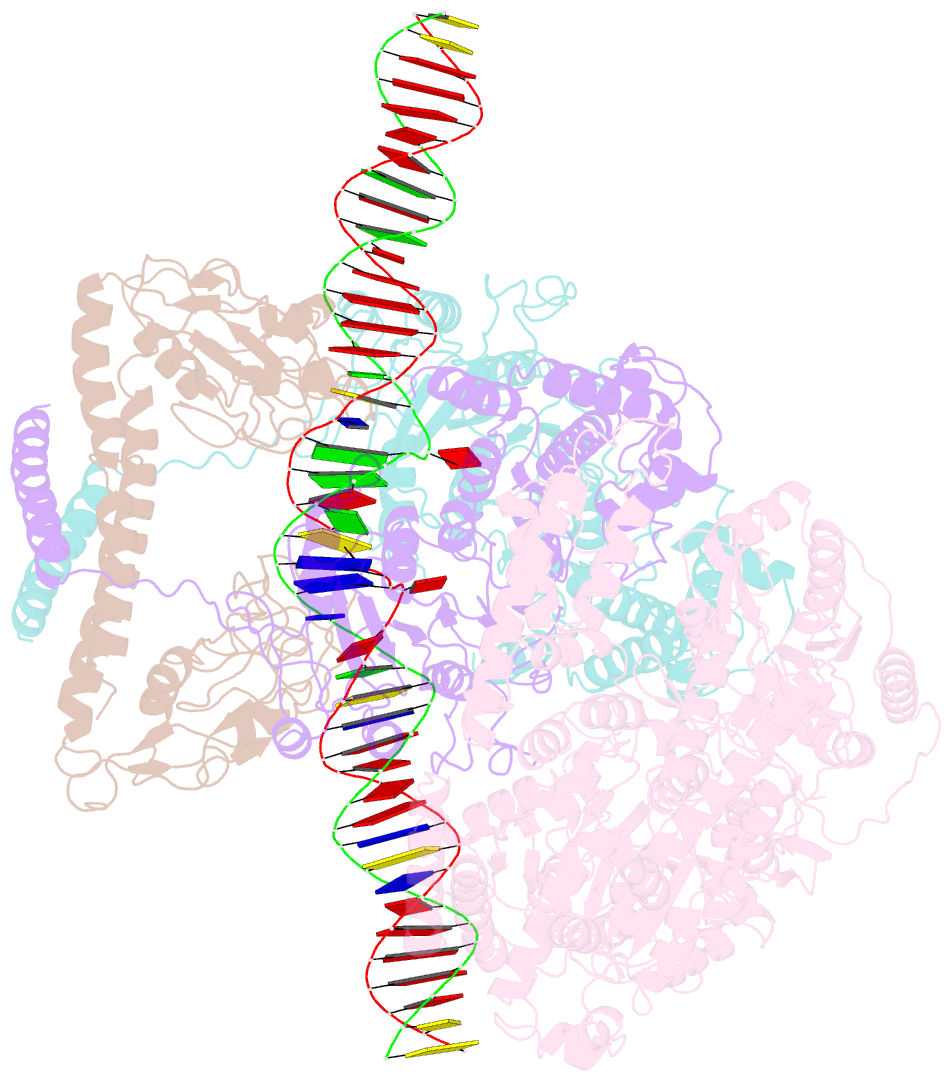
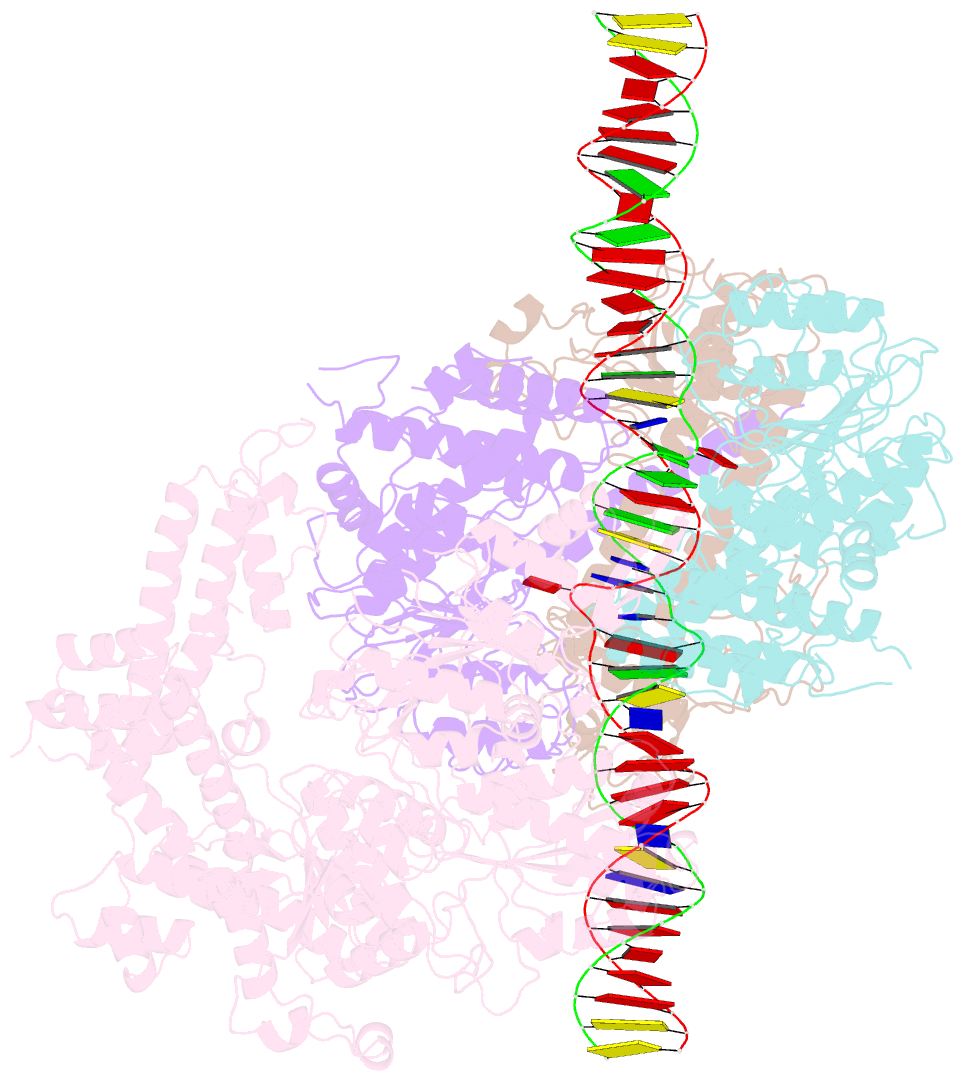
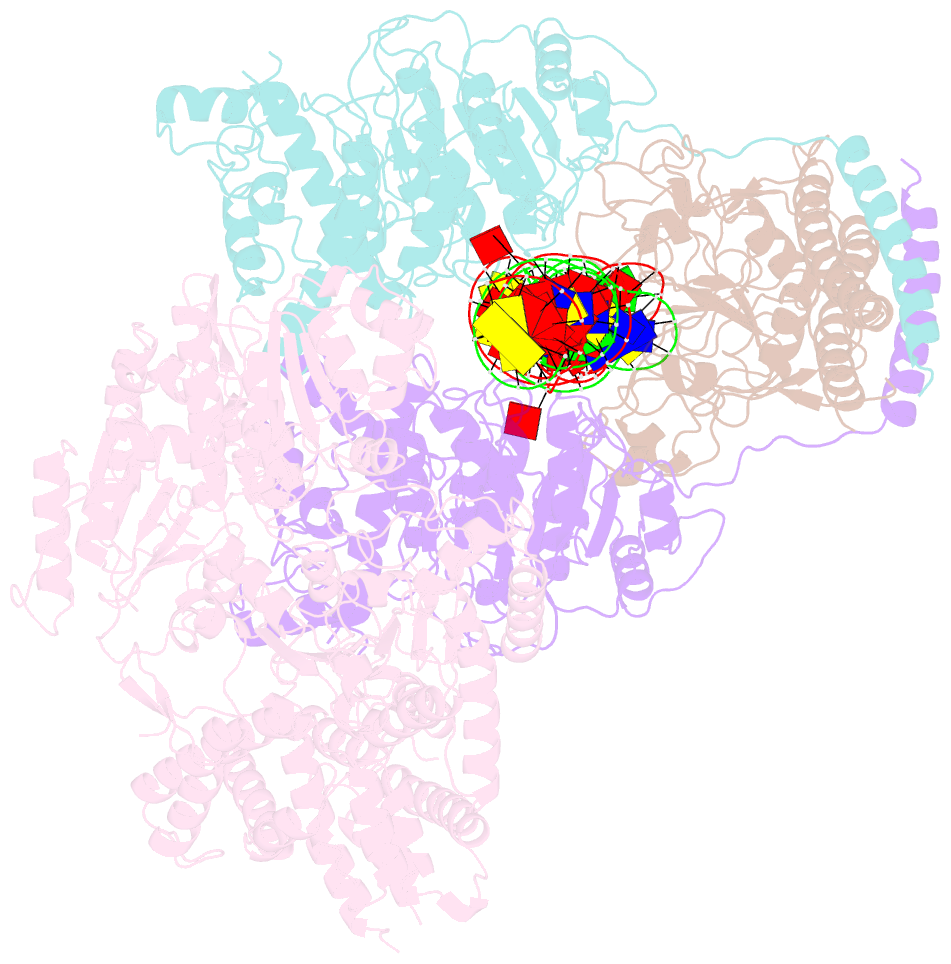
Summary information and primary citation
- PDB-id
- 7btq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- cryo-EM (4.54 Å)
- Summary
- Ecor124i-DNA in the restriction-alleviation state
- Reference
- Gao Y, Cao D, Zhu J, Feng H, Luo X, Liu S, Yan XX, Zhang X, Gao P (2020): "Structural insights into assembly, operation and inhibition of a type I restriction-modification system." Nat Microbiol, 5, 1107-1118. doi: 10.1038/s41564-020-0731-z.
- Abstract
- Type I restriction-modification (R-M) systems are widespread in prokaryotic genomes and provide robust protection against foreign DNA. They are multisubunit enzymes with methyltransferase, endonuclease and translocase activities. Despite extensive studies over the past five decades, little is known about the molecular mechanisms of these sophisticated machines. Here, we report the cryo-electron microscopy structures of the representative EcoR124I R-M system in different assemblies (R2M2S1, R1M2S1 and M2S1) bound to target DNA and the phage and mobile genetic element-encoded anti-restriction proteins Ocr and ArdA. EcoR124I can precisely regulate different enzymatic activities by adopting distinct conformations. The marked conformational transitions of EcoR124I are dependent on the intrinsic flexibility at both the individual-subunit and assembled-complex levels. Moreover, Ocr and ArdA use a DNA-mimicry strategy to inhibit multiple activities, but do not block the conformational transitions of the complexes. These structural findings, complemented by mutational studies of key intermolecular contacts, provide insights into assembly, operation and inhibition mechanisms of type I R-M systems.