Summary information and primary citation
- PDB-id
- 7bv2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (2.5 Å)
- Summary
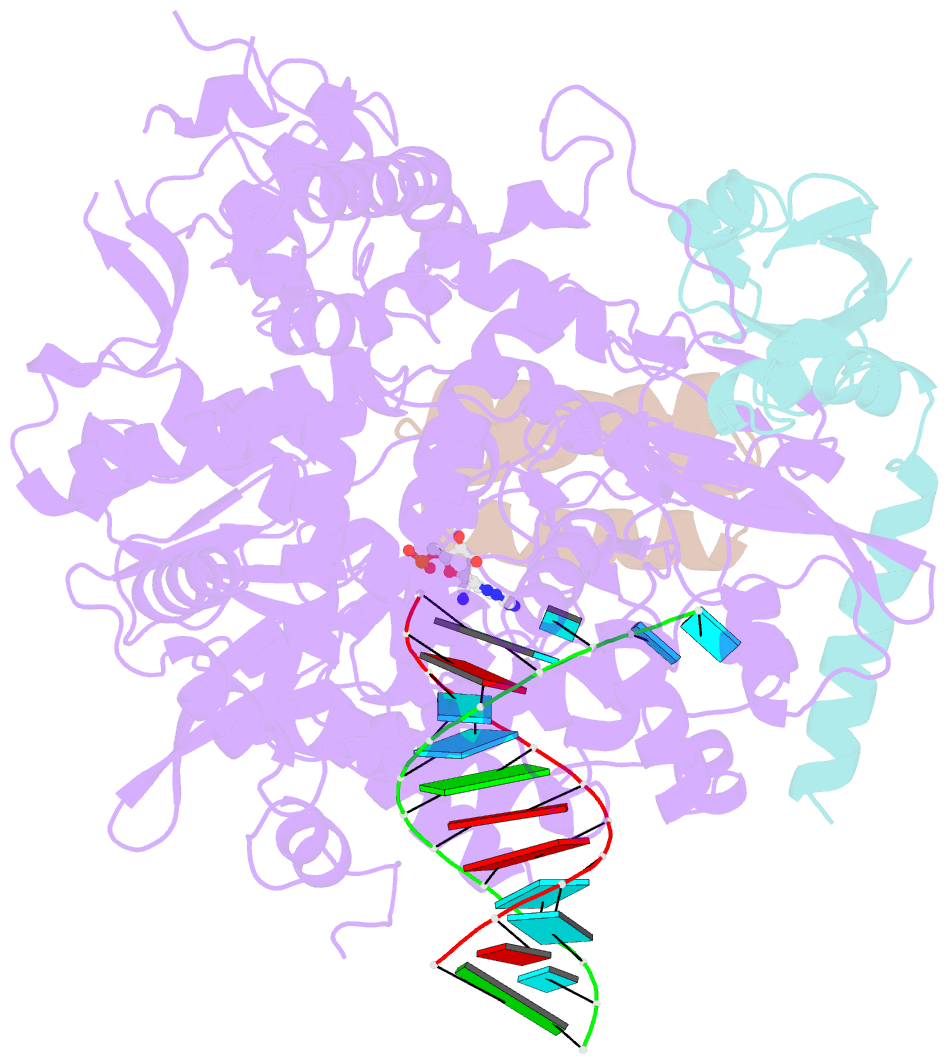
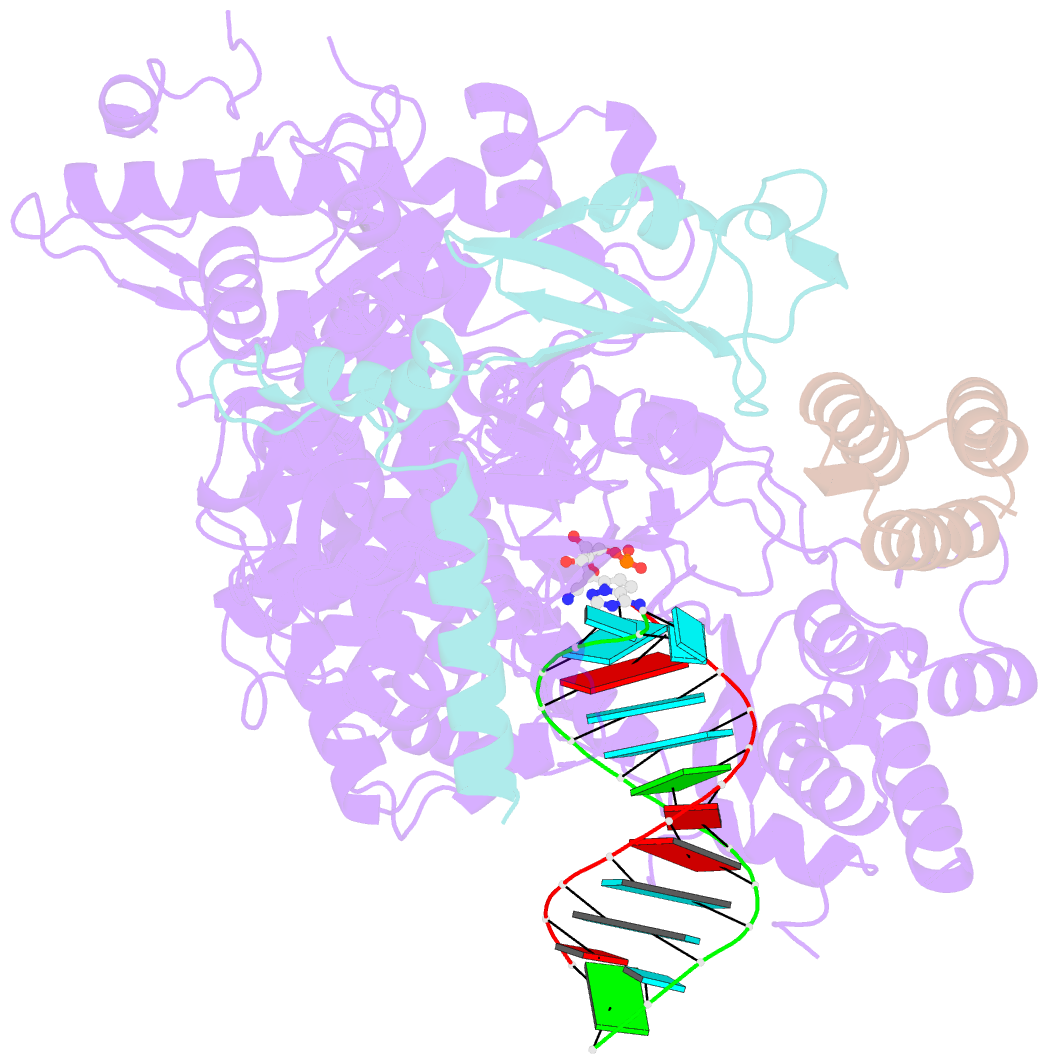
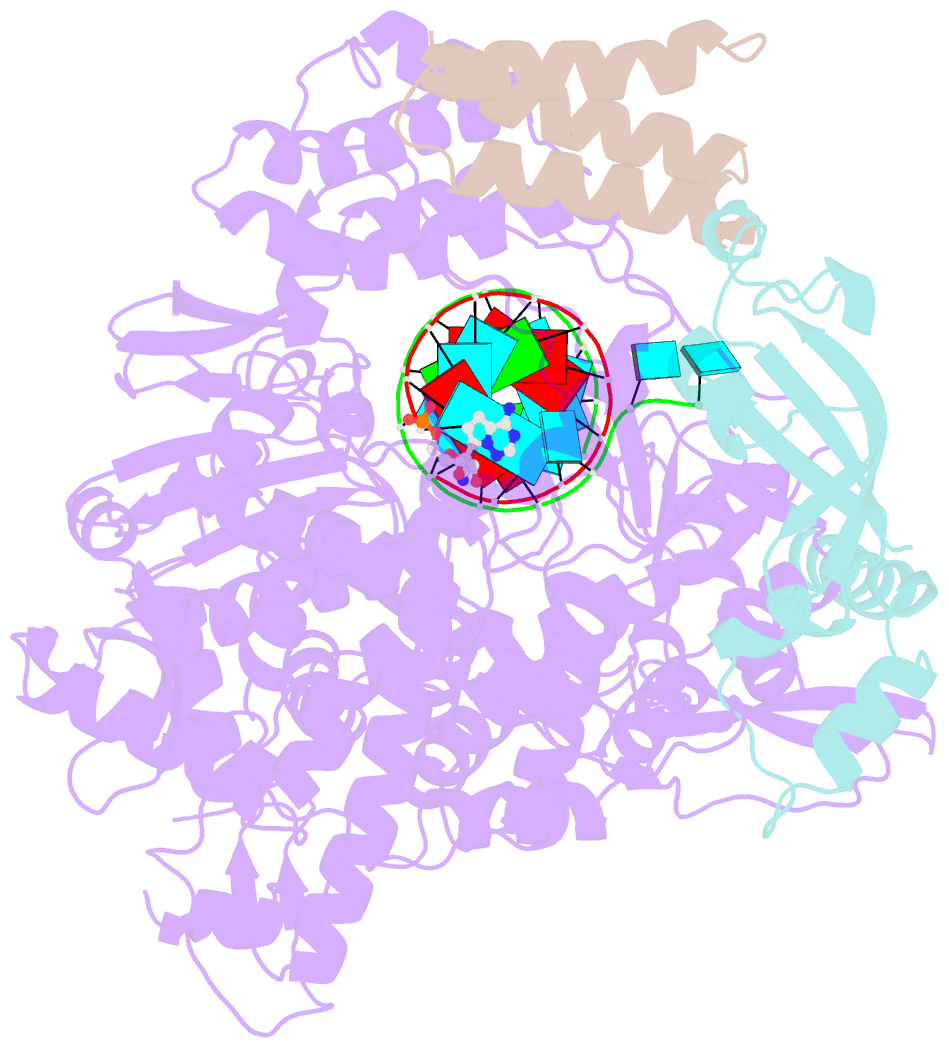
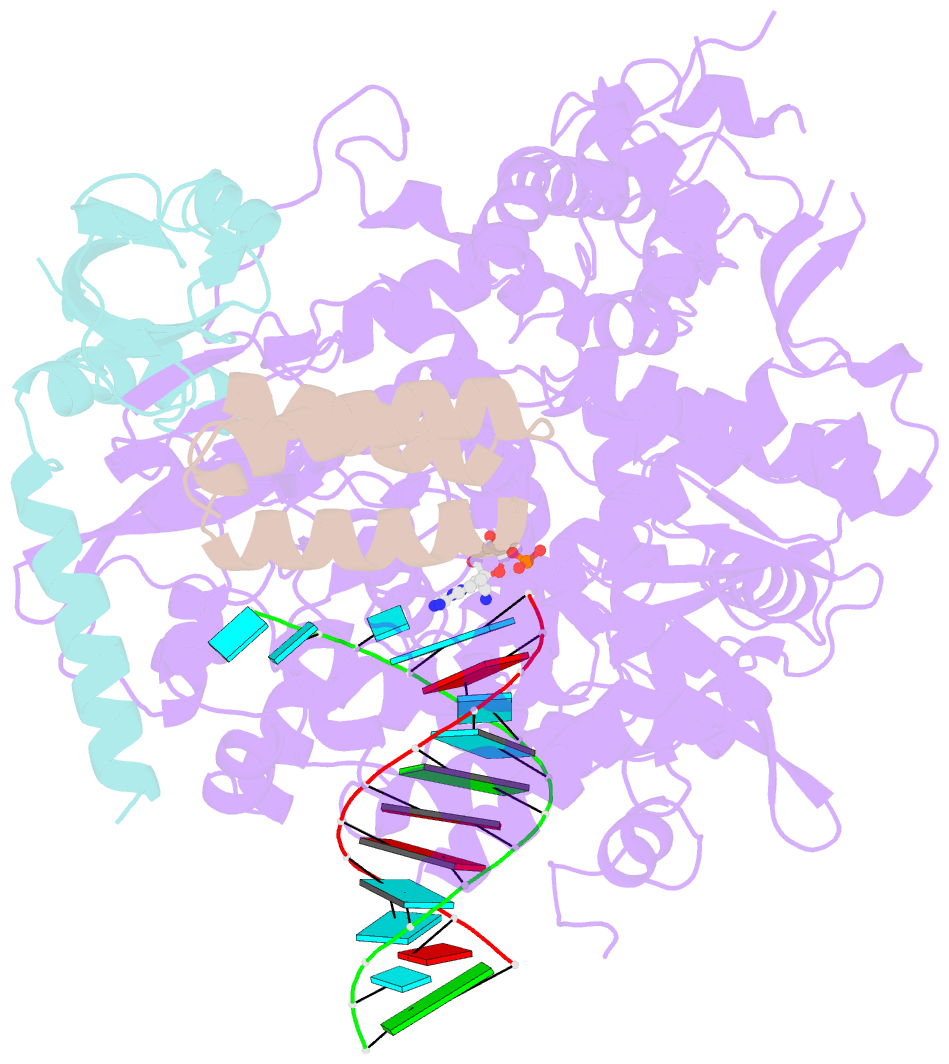
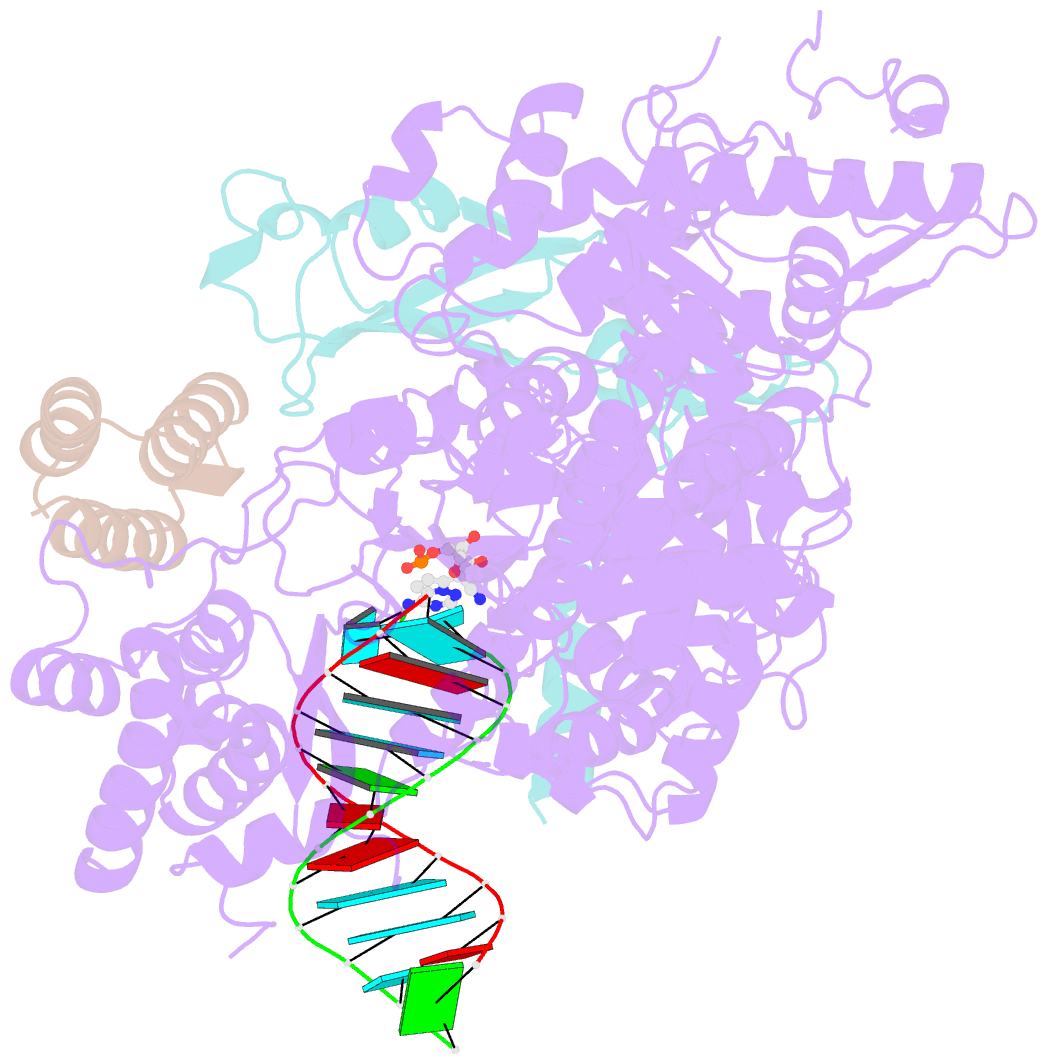
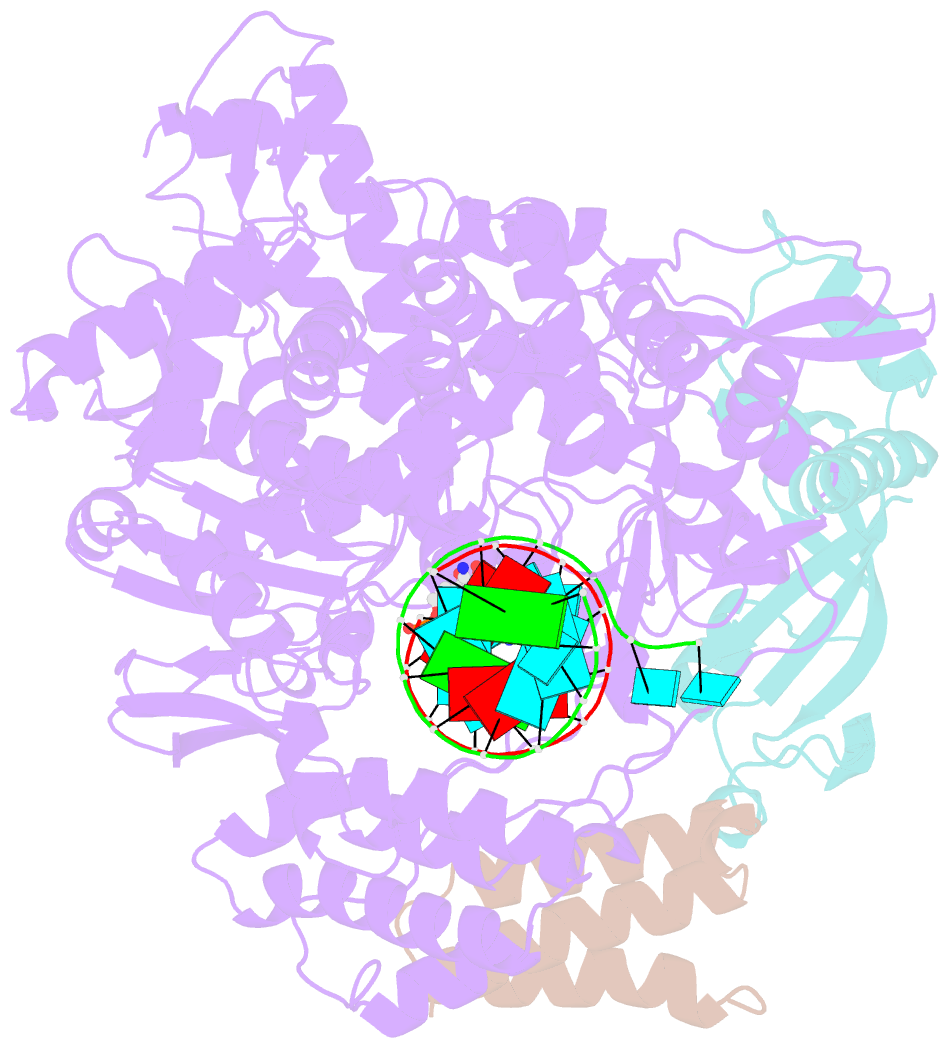
- The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of remdesivir(rtp)
- Reference
- Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE (2020): "Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir." Science, 368, 1499-1504. doi: 10.1126/science.abc1560.
- Abstract
- The pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global crisis. Replication of SARS-CoV-2 requires the viral RNA-dependent RNA polymerase (RdRp) enzyme, a target of the antiviral drug remdesivir. Here we report the cryo-electron microscopy structure of the SARS-CoV-2 RdRp, both in the apo form at 2.8-angstrom resolution and in complex with a 50-base template-primer RNA and remdesivir at 2.5-angstrom resolution. The complex structure reveals that the partial double-stranded RNA template is inserted into the central channel of the RdRp, where remdesivir is covalently incorporated into the primer strand at the first replicated base pair, and terminates chain elongation. Our structures provide insights into the mechanism of viral RNA replication and a rational template for drug design to combat the viral infection.