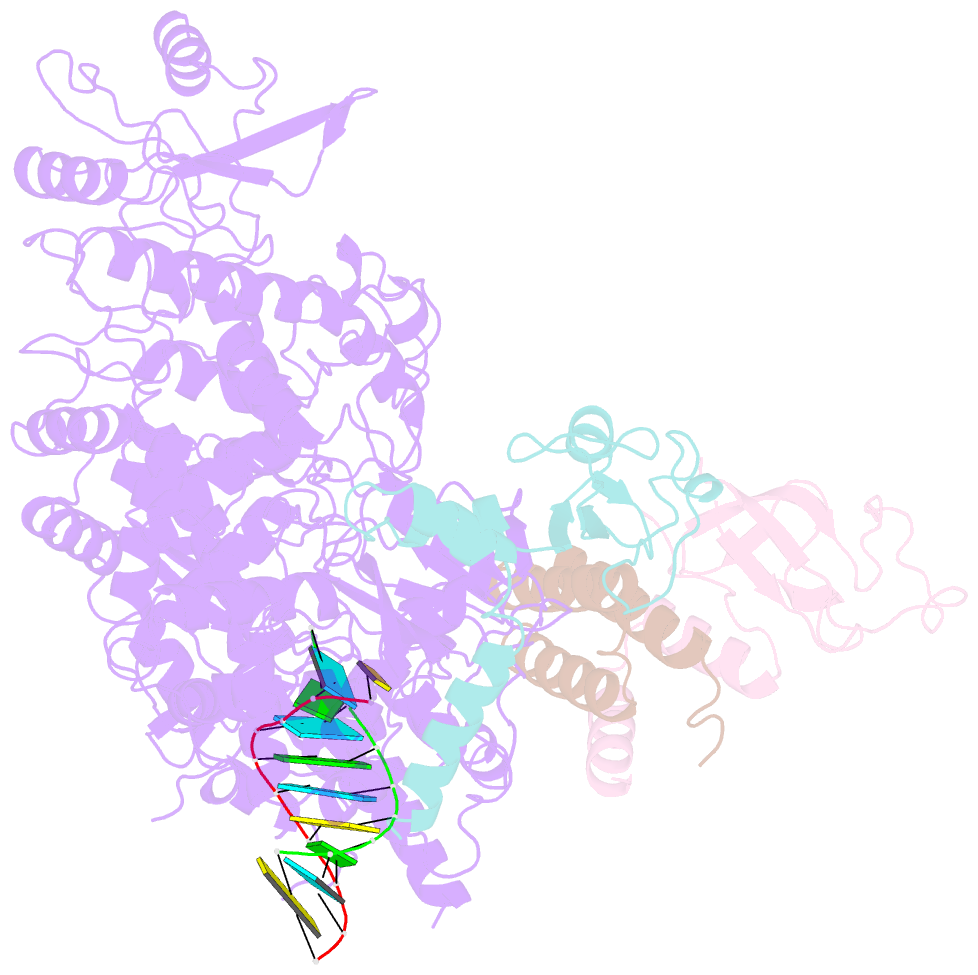
Summary information and primary citation
- PDB-id
- 7bzf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- cryo-EM (3.26 Å)
- Summary
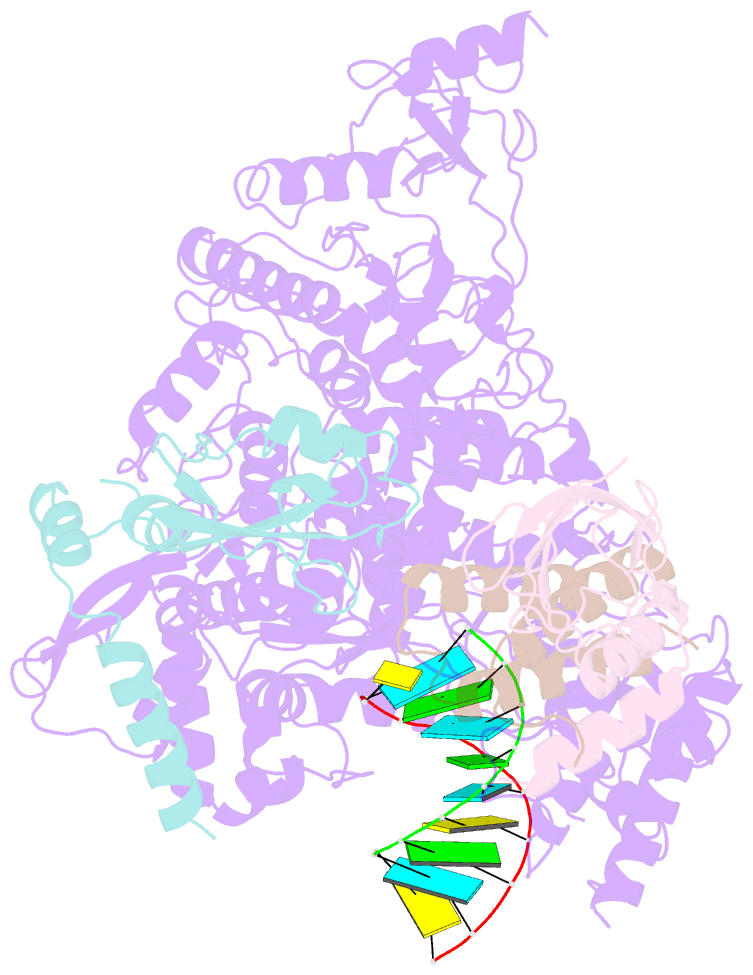
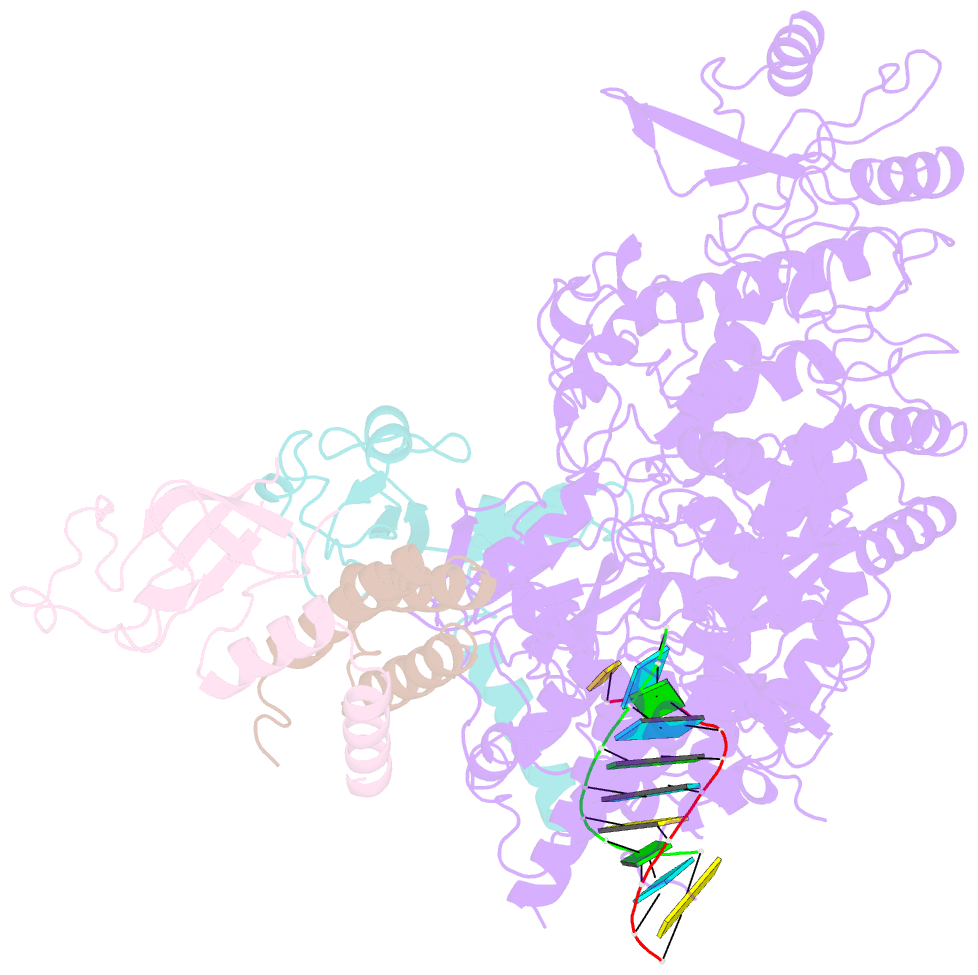
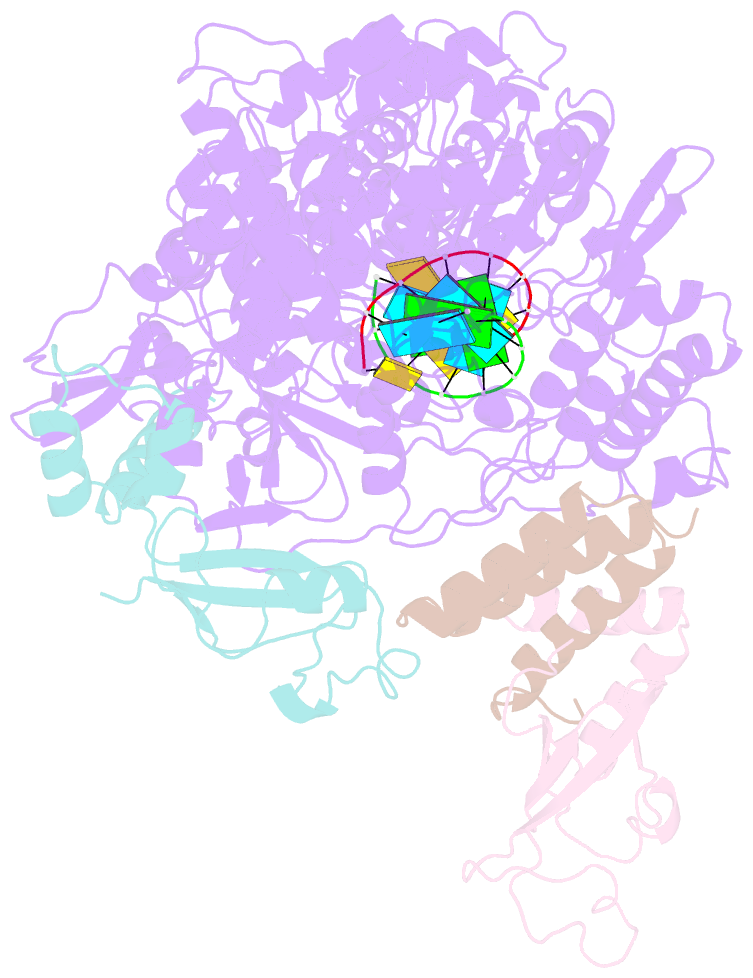
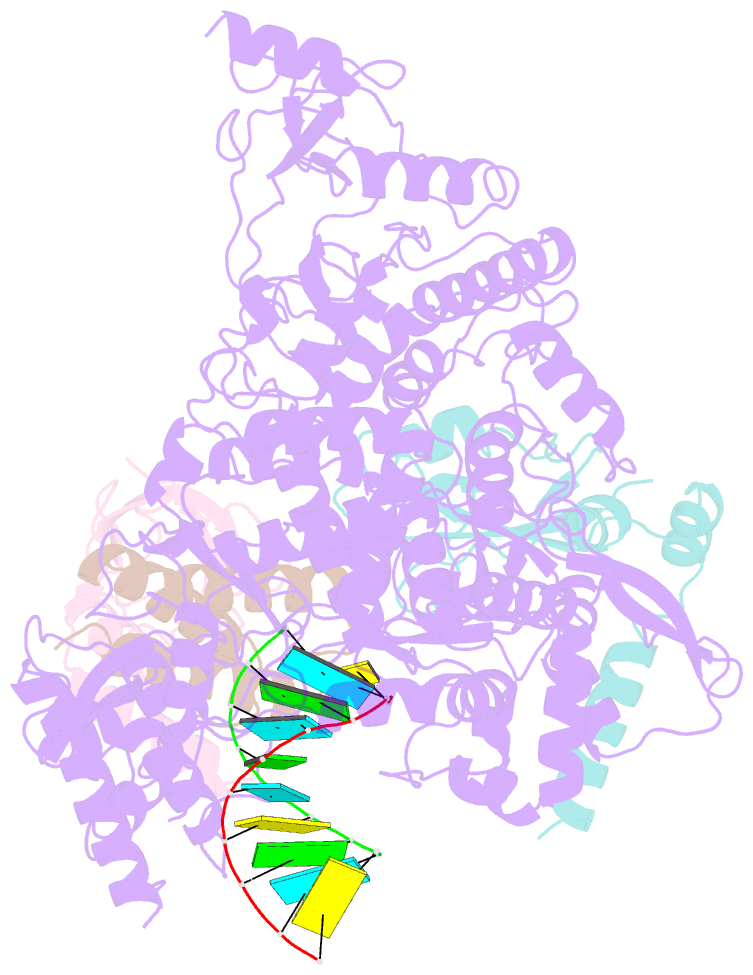
- Covid-19 RNA-dependent RNA polymerase post-translocated catalytic complex
- Reference
- Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, Fang X, Yang X, Huang Y, Gao H, Liu F, Ge J, Sun Q, Yang X, Xu W, Liu Z, Yang H, Lou Z, Jiang B, Guddat LW, Gong P, Rao Z (2020): "Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase." Cell, 182, 417-428.e13. doi: 10.1016/j.cell.2020.05.034.
- Abstract
- Nucleotide analog inhibitors, including broad-spectrum remdesivir and favipiravir, have shown promise in in vitro assays and some clinical studies for COVID-19 treatment, this despite an incomplete mechanistic understanding of the viral RNA-dependent RNA polymerase nsp12 drug interactions. Here, we examine the molecular basis of SARS-CoV-2 RNA replication by determining the cryo-EM structures of the stalled pre- and post- translocated polymerase complexes. Compared with the apo complex, the structures show notable structural rearrangements happening to nsp12 and its co-factors nsp7 and nsp8 to accommodate the nucleic acid, whereas there are highly conserved residues in nsp12, positioning the template and primer for an in-line attack on the incoming nucleotide. Furthermore, we investigate the inhibition mechanism of the triphosphate metabolite of remdesivir through structural and kinetic analyses. A transition model from the nsp7-nsp8 hexadecameric primase complex to the nsp12-nsp7-nsp8 polymerase complex is also proposed to provide clues for the understanding of the coronavirus transcription and replication machinery.