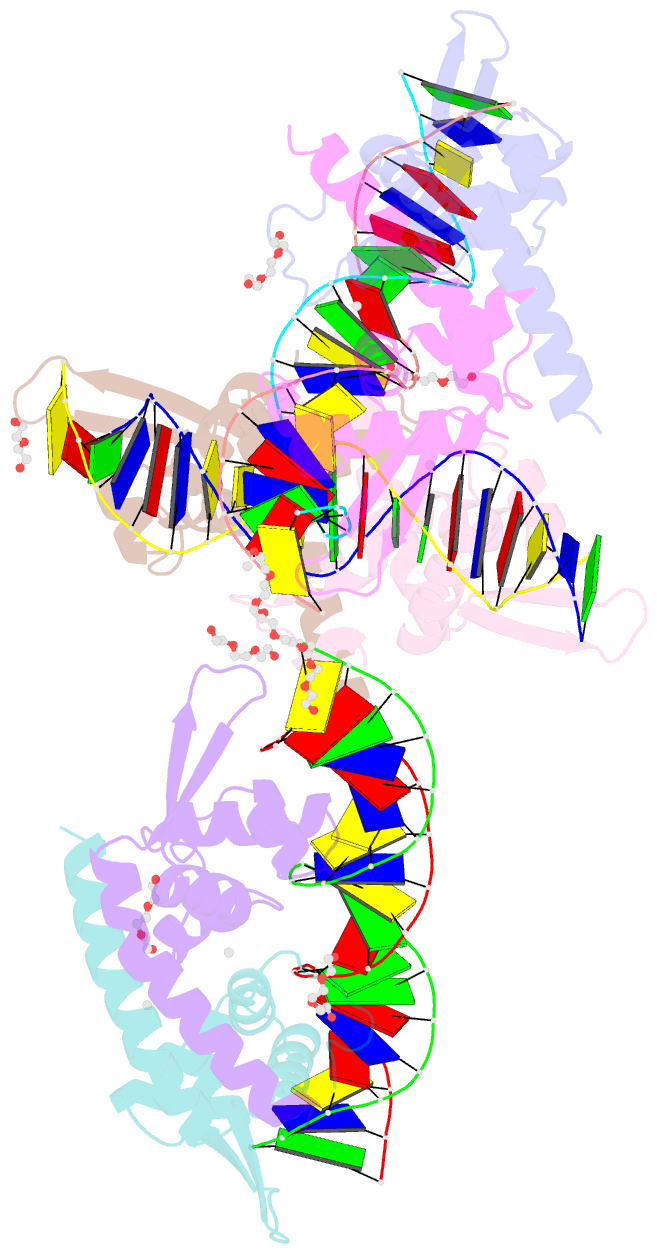
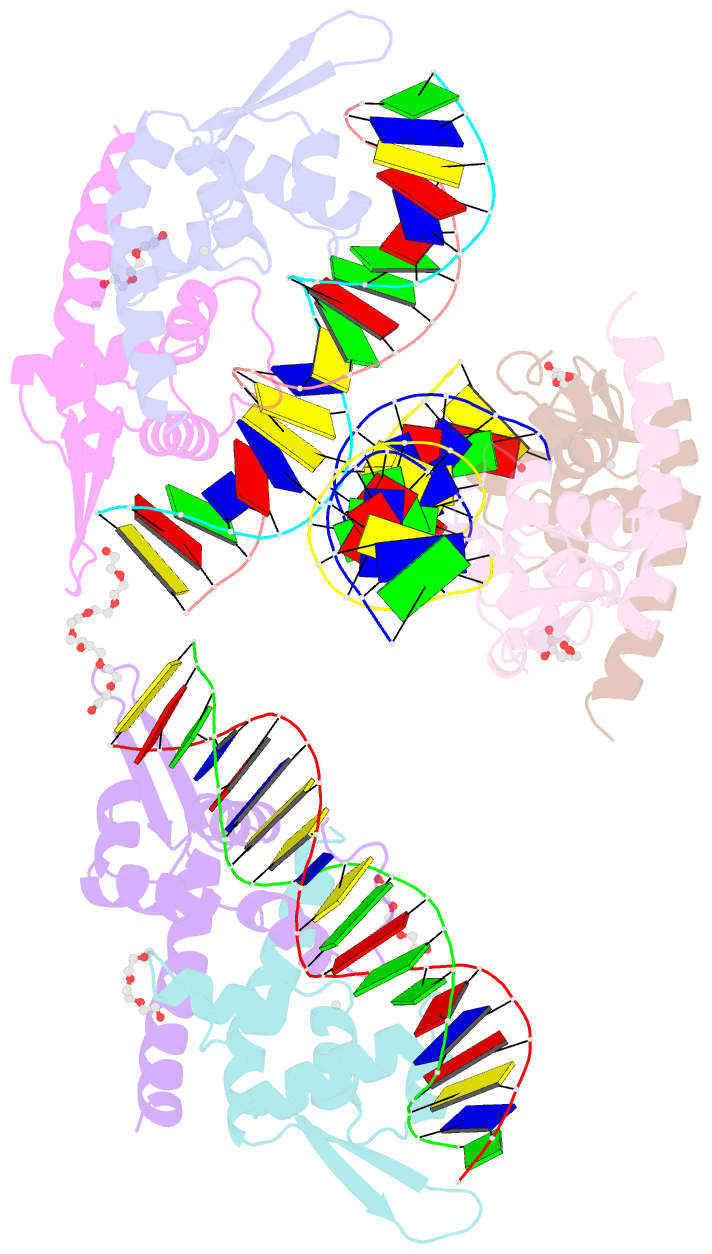
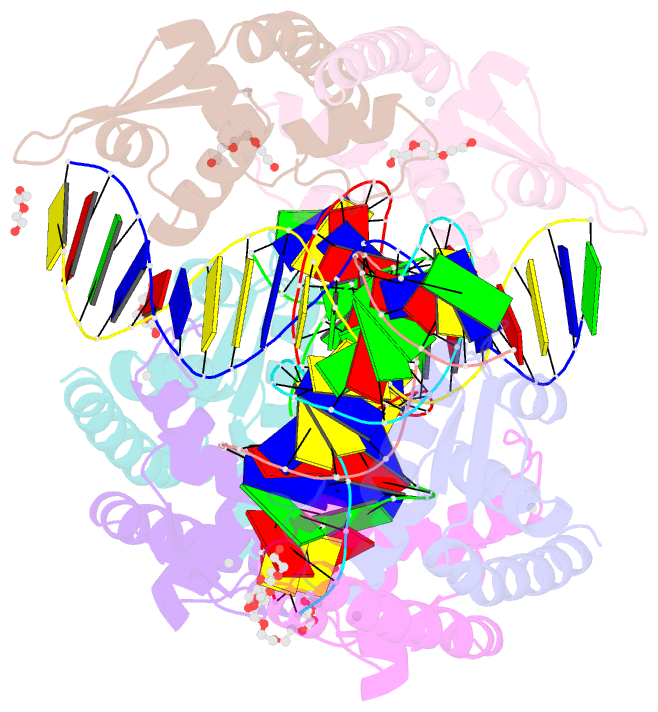
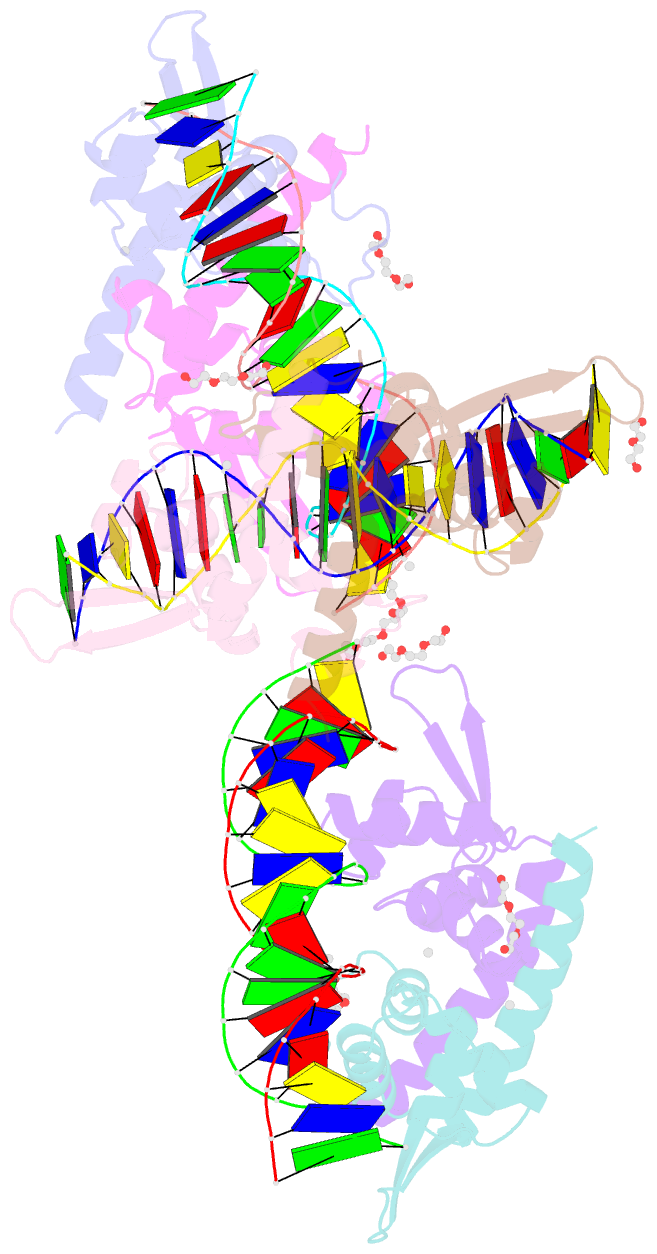
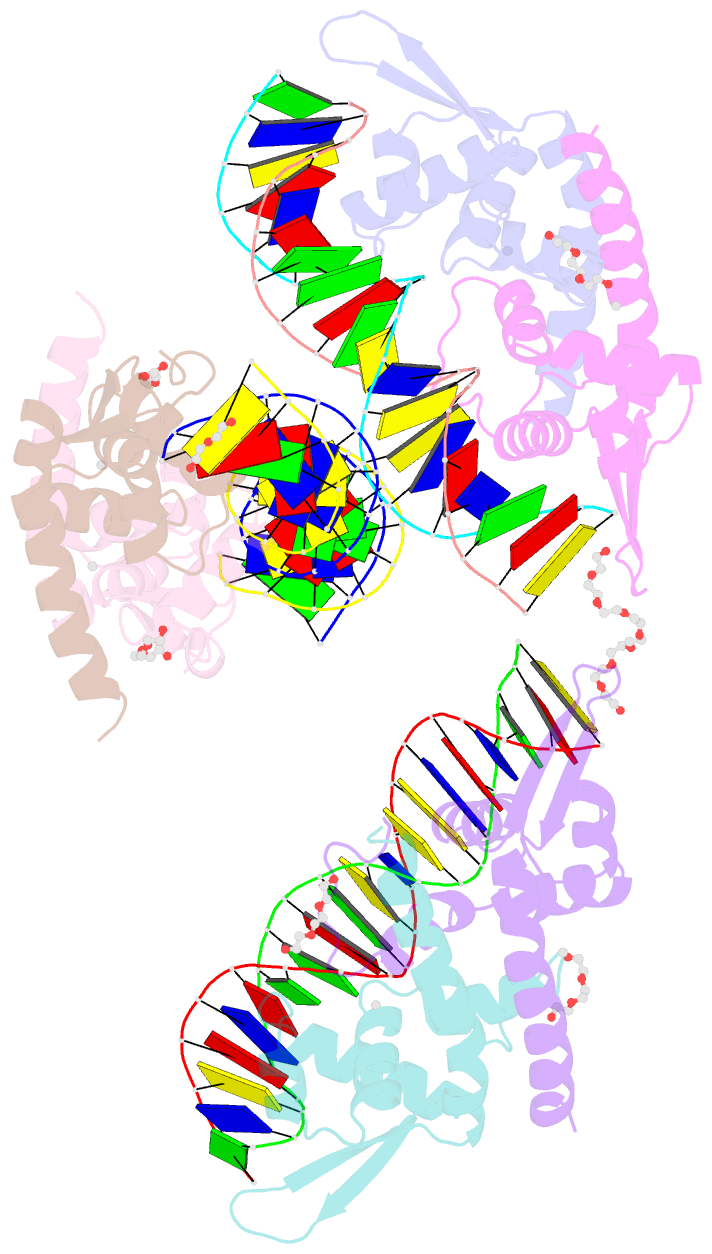
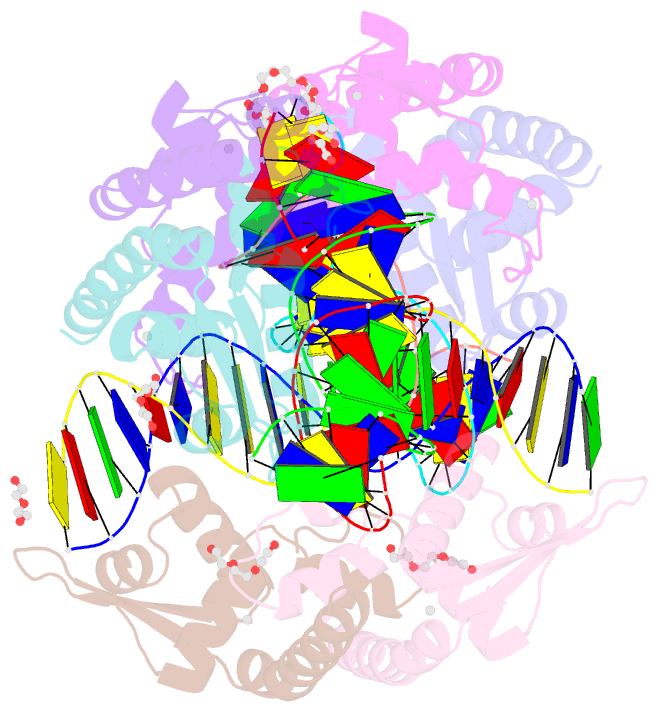
Summary information and primary citation
- PDB-id
- 7bzg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.9 Å)
- Summary
- Structure of bacillus subtilis hxlr, wild type in complex with formaldehyde and DNA
- Reference
- Zhu R, Zhang G, Jing M, Han Y, Li J, Zhao J, Li Y, Chen PR (2021): "Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction." Nat Commun, 12, 581. doi: 10.1038/s41467-020-20754-4.
- Abstract
- Formaldehyde (FA) has long been considered as a toxin and carcinogen due to its damaging effects to biological macromolecules, but its beneficial roles have been increasingly appreciated lately. Real-time monitoring of this reactive molecule in living systems is highly desired in order to decipher its physiological and/or pathological functions, but a genetically encoded FA sensor is currently lacking. We herein adopt a structure-based study of the underlying mechanism of the FA-responsive transcription factor HxlR from Bacillus subtilis, which shows that HxlR recognizes FA through an intra-helical cysteine-lysine crosslinking reaction at its N-terminal helix α1, leading to conformational change and transcriptional activation. By leveraging this FA-induced intra-helical crosslinking and gain-of-function reorganization, we develop the genetically encoded, reaction-based FA sensor-FAsor, allowing spatial-temporal visualization of FA in mammalian cells and mouse brain tissues.