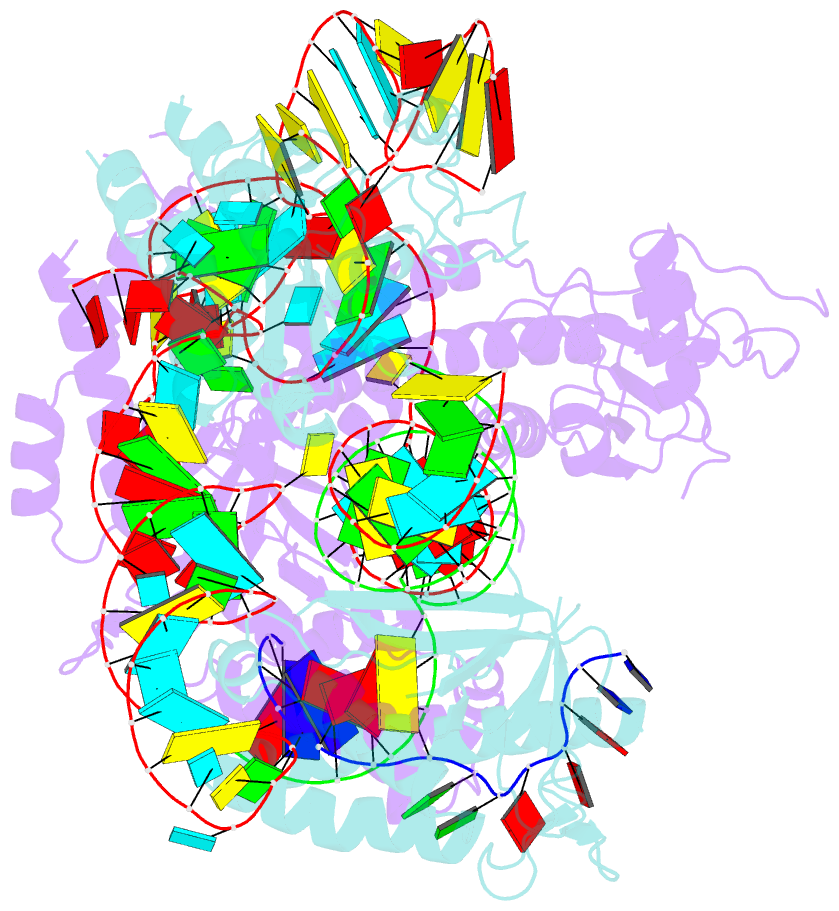
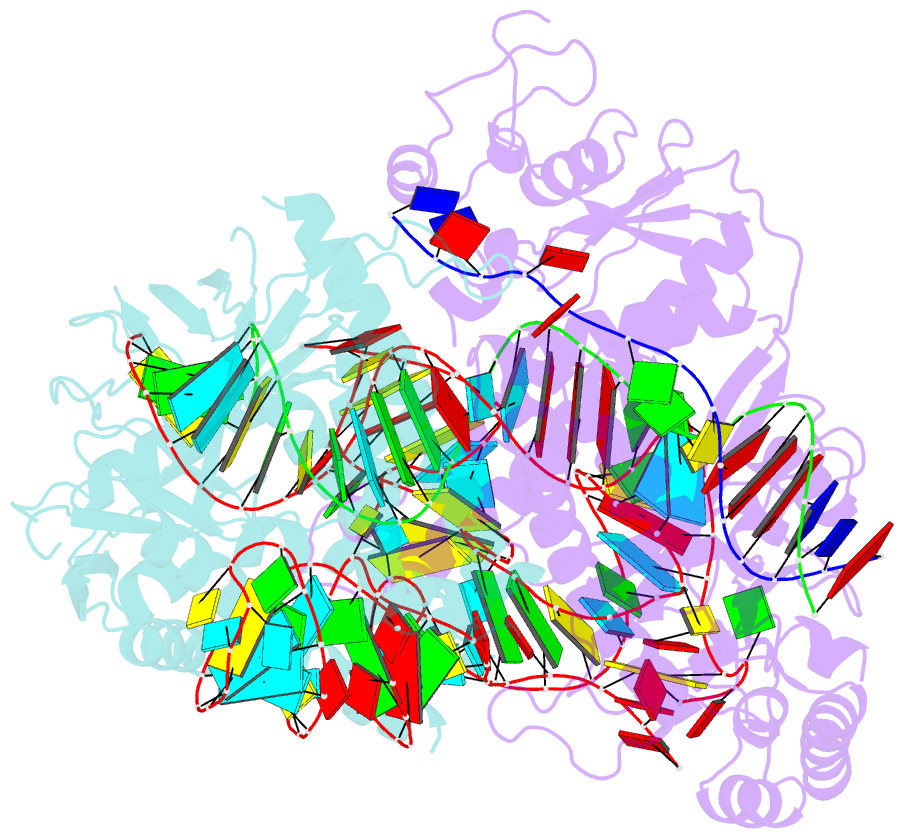
Summary information and primary citation
- PDB-id
- 7c7l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (3.3 Å)
- Summary
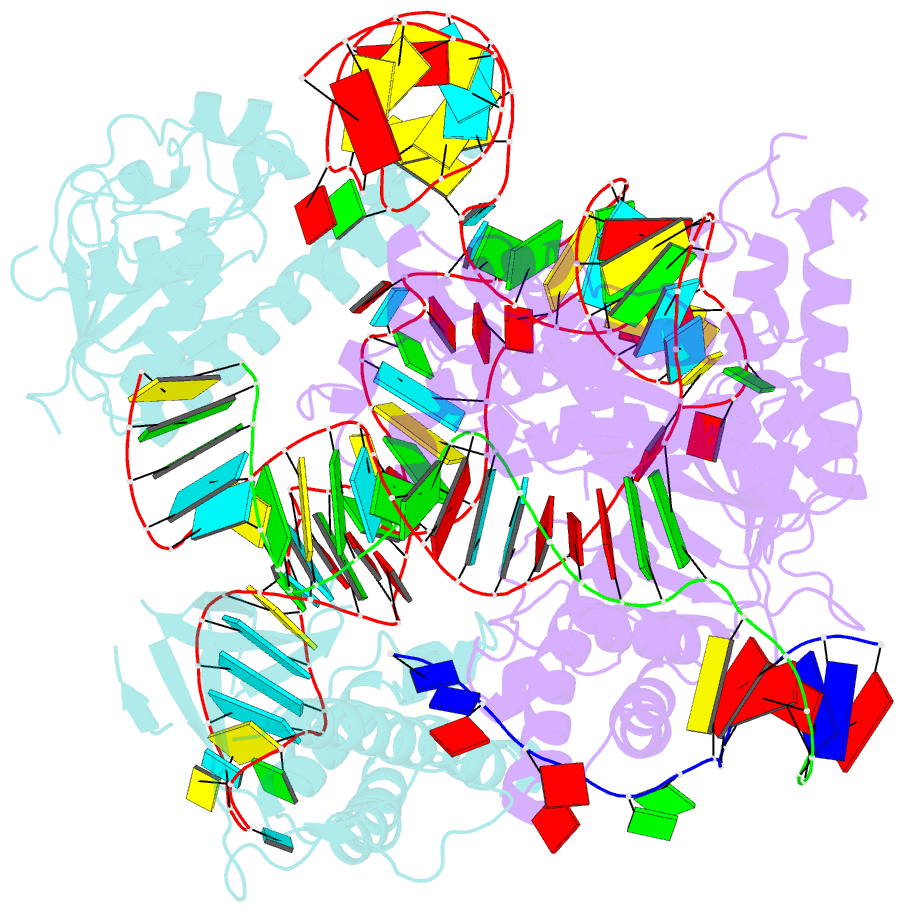
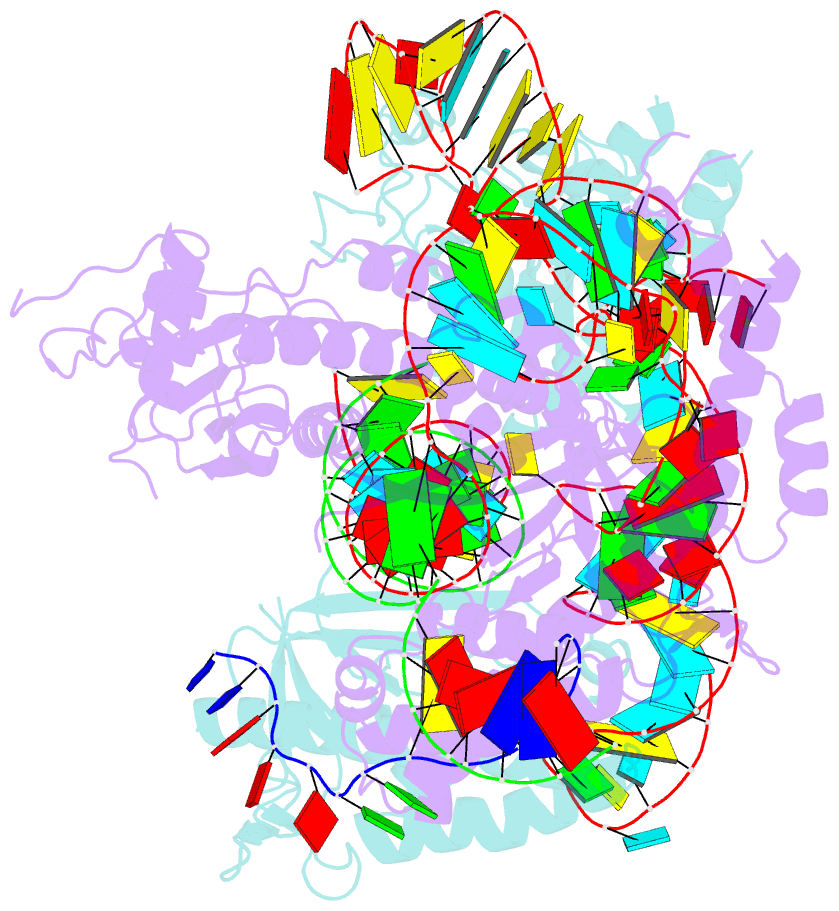
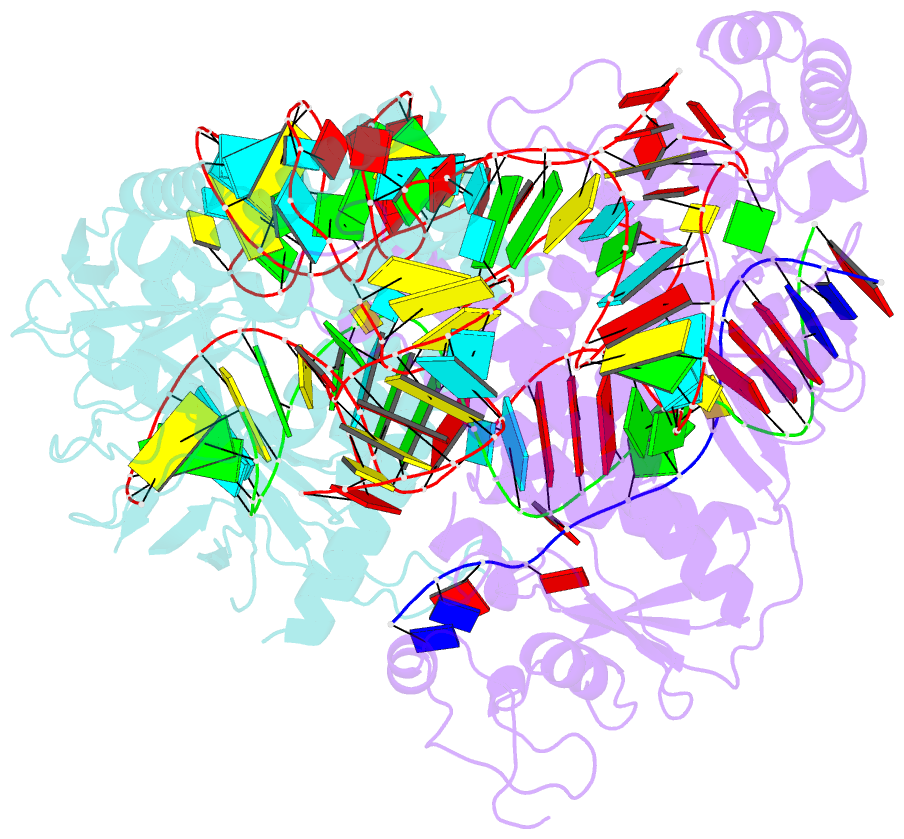
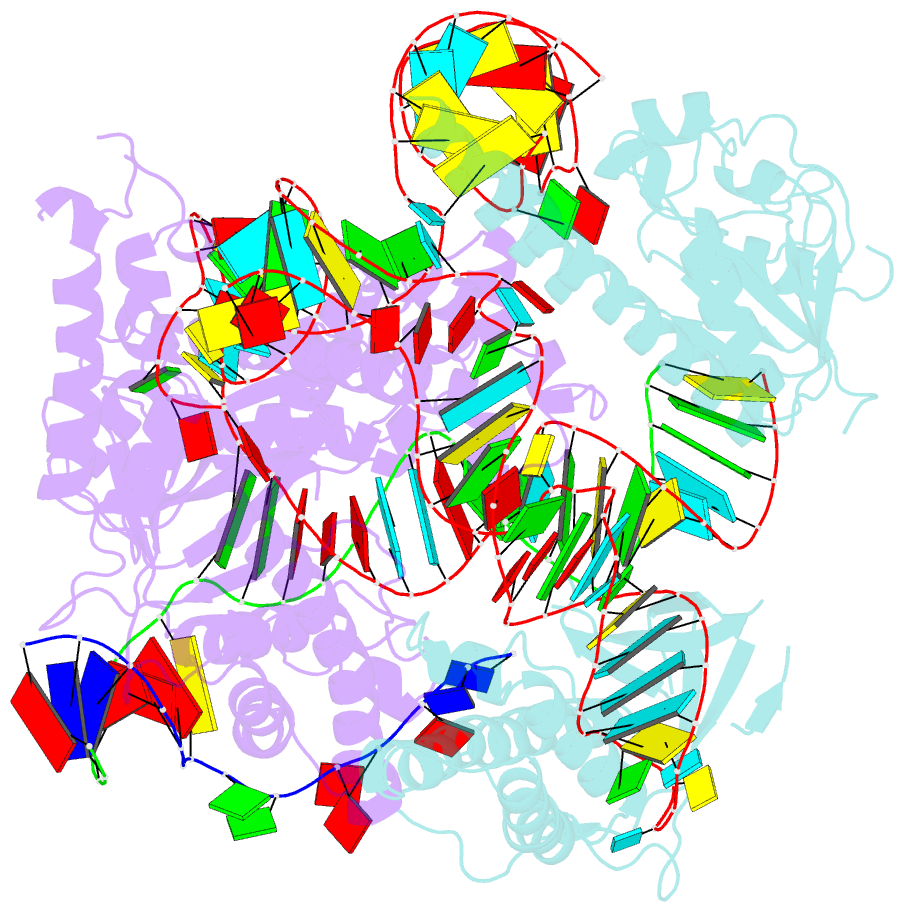
- cryo-EM structure of the cas12f1-sgrna-target DNA complex
- Reference
- Takeda SN, Nakagawa R, Okazaki S, Hirano H, Kobayashi K, Kusakizako T, Nishizawa T, Yamashita K, Nishimasu H, Nureki O (2021): "Structure of the miniature type V-F CRISPR-Cas effector enzyme." Mol.Cell, 81, 558-570.e3. doi: 10.1016/j.molcel.2020.11.035.
- Abstract
- RNA-guided DNA endonucleases derived from CRISPR-Cas adaptive immune systems are widely used as powerful genome-engineering tools. Among the diverse CRISPR-Cas nucleases, the type V-F Cas12f (also known as Cas14) proteins are exceptionally compact and associate with a guide RNA to cleave single- and double-stranded DNA targets. Here, we report the cryo-electron microscopy structure of Cas12f1 (also known as Cas14a) in complex with a guide RNA and its target DNA. Unexpectedly, the structure revealed that two Cas12f1 molecules assemble with the single guide RNA to recognize the double-stranded DNA target. Each Cas12f1 protomer adopts a different conformation and plays distinct roles in nucleic acid recognition and DNA cleavage, thereby explaining how the miniature Cas12f1 enzyme achieves RNA-guided DNA cleavage as an "asymmetric homodimer." Our findings augment the mechanistic understanding of diverse CRISPR-Cas nucleases and provide a framework for the development of compact genome-engineering tools critical for therapeutic genome editing.