Summary information and primary citation
- PDB-id
- 7c9o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.55 Å)
- Summary
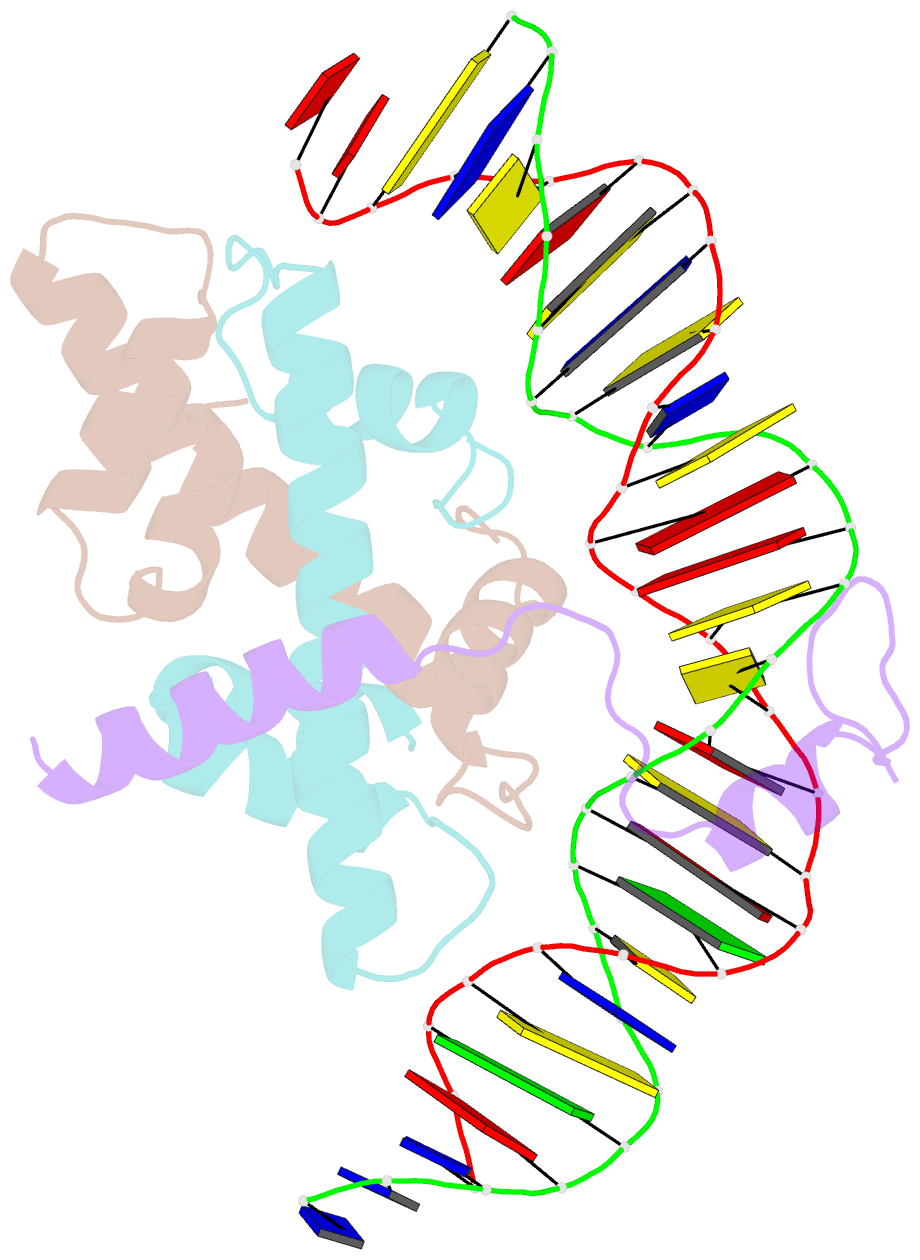
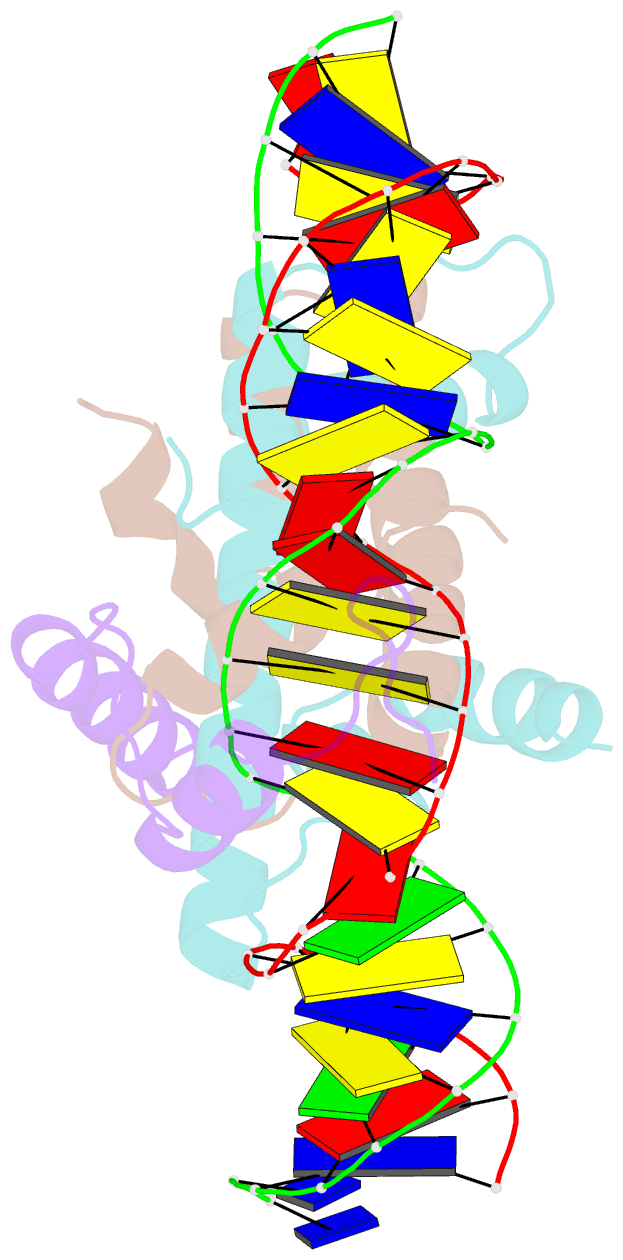
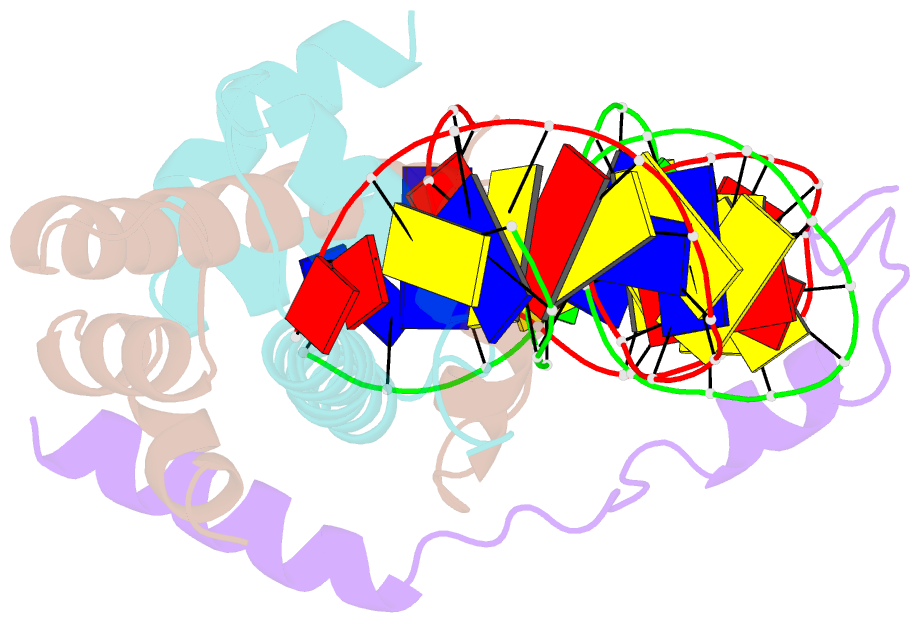
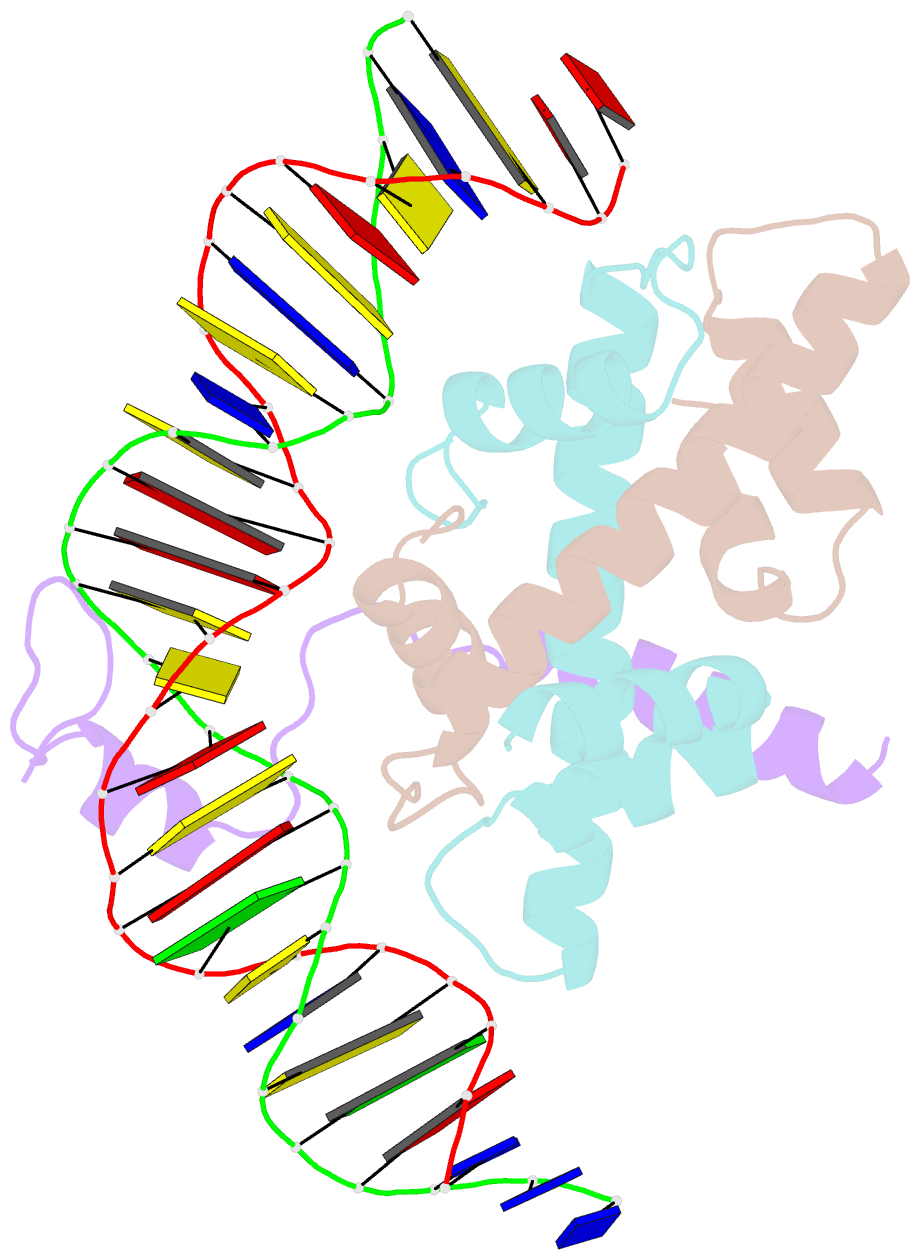
- Crystal structure of DNA-bound cct-nf-yb-yc complex (hd1cct-ghd8-osnf-yc2)
- Reference
- Shen C, Liu H, Guan Z, Yan J, Zheng T, Yan W, Wu C, Zhang Q, Yin P, Xing Y (2020): "Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering." Plant Cell, 32, 3469-3484. doi: 10.1105/tpc.20.00067.
- Abstract
- CONSTANS, CONSTANS-LIKE, and TIMING OF CAB EXPRESSION1 (CCT) domain-containing proteins are a large family unique to plants. They transcriptionally regulate photoperiodic flowering, circadian rhythms, vernalization, and other related processes. Through their CCT domains, CONSTANS and HEADING DATE1 (HD1) coordinate with the NUCLEAR FACTOR Y (NF-Y) B/C dimer to specifically target a conserved 'CCACA' motif within the promoters of their target genes. However, the mechanism underlying DNA recognition by the CCT domain remains unclear. Here we determined the crystal structures of the rice (Oryza sativa) NF-YB/YC dimer and the florigen gene Heading date 3a (Hd3a)-bound HD1CCT/NF-YB/YC trimer with resolutions of 2.0 Å and 2.55 Å, respectively. The CCT domain of HD1 displays an elongated structure containing two α-helices and two loops, tethering Hd3a to the NF-YB/YC dimer. Helix α2 and loop 2 are anchored into the minor groove of the 'CCACA' motif, which determines the specific base recognition. Our structures reveal the interaction mechanism among the CCT domain, NF-YB/YC dimer, and the target DNA. These results not only provide insight into the network between the CCT proteins and NF-Y subunits, but also offer potential approaches for improving productivity and global adaptability of crops by manipulating florigen expression.