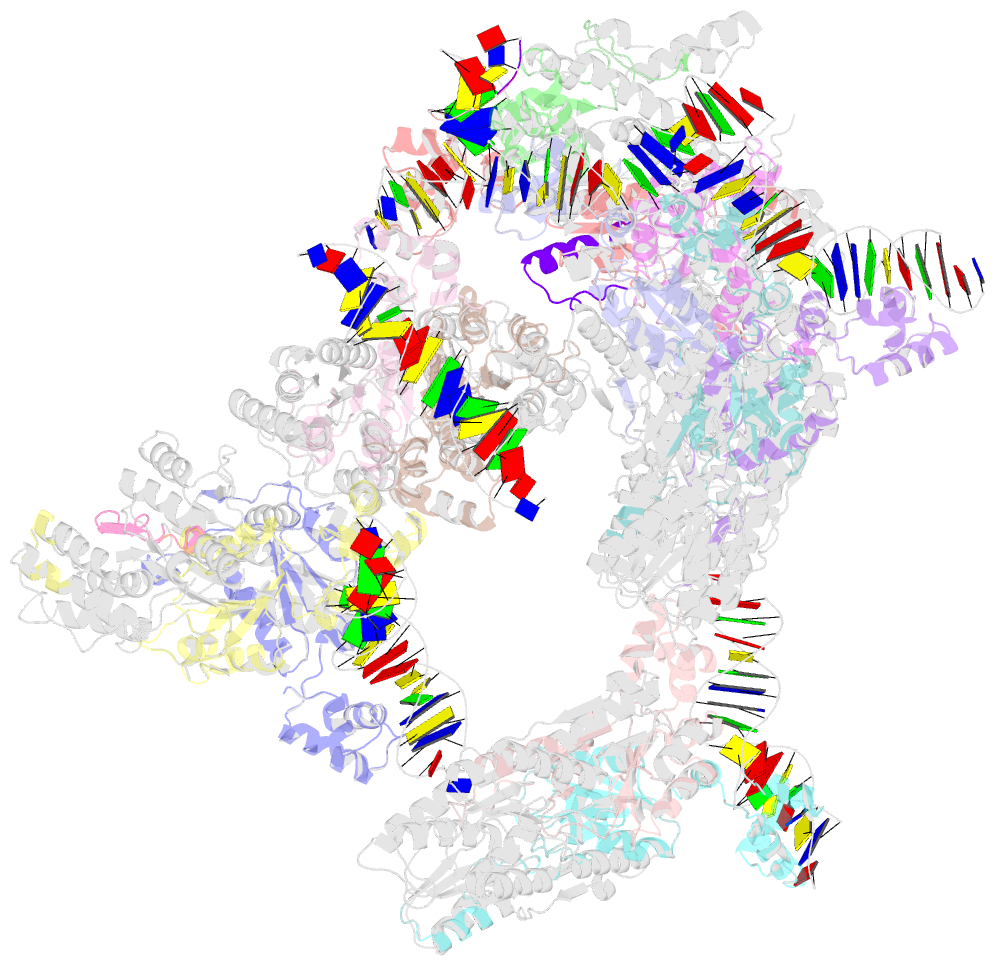
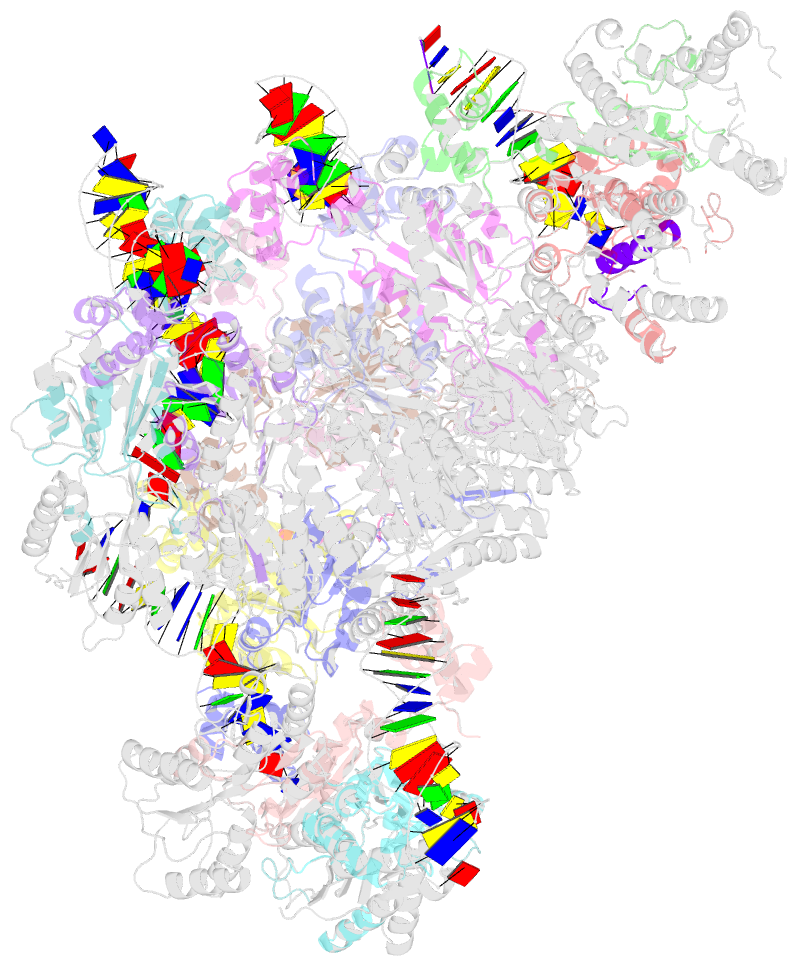
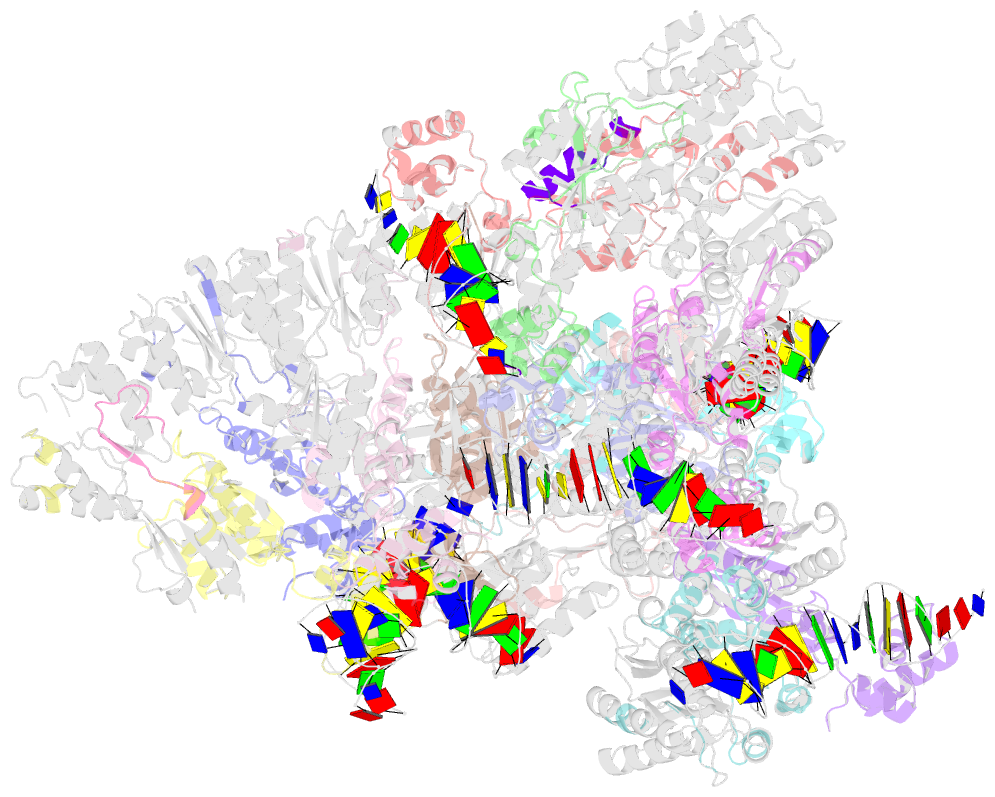
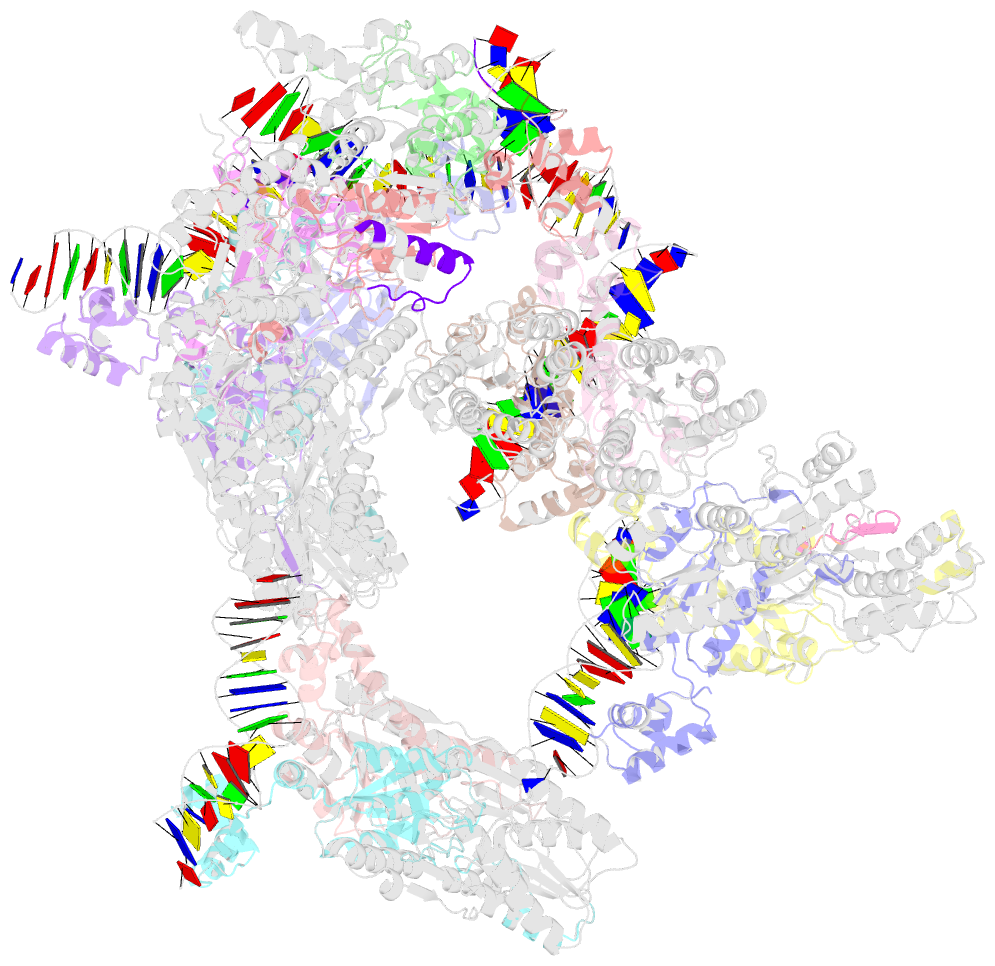
Summary information and primary citation
- PDB-id
- 7ce1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.2 Å)
- Summary
- Complex structure of transcription factor sghr with its cognate DNA
- Reference
- Ye F, Wang C, Fu Q, Yan XF, Bharath SR, Casanas A, Wang M, Song H, Zhang LH, Gao YG (2020): "Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants." J.Biol.Chem., 295, 12290-12304. doi: 10.1074/jbc.RA120.012908.
- Abstract
- Agrobacterium tumefaciens infects various plants and causes crown gall diseases involving temporal expression of virulence factors. SghA is a newly identified virulence factor enzymatically releasing salicylic acid from its glucoside conjugate and controlling plant tumor development. Here, we report the structural basis of SghR, a LacI-type transcription factor highly conserved in Rhizobiaceae family, regulating the expression of SghA and involved in tumorigenesis. We identified and characterized the binding site of SghR on the promoter region of sghA and then determined the crystal structures of apo-SghR, SghR complexed with its operator DNA, and ligand sucrose, respectively. These results provide detailed insights into how SghR recognizes its cognate DNA and shed a mechanistic light on how sucrose attenuates the affinity of SghR with DNA to modulate the expression of SghA. Given the important role of SghR in mediating the signaling cross-talk during Agrobacterium infection, our results pave the way for structure-based inducer analog design, which has potential applications for agricultural industry.