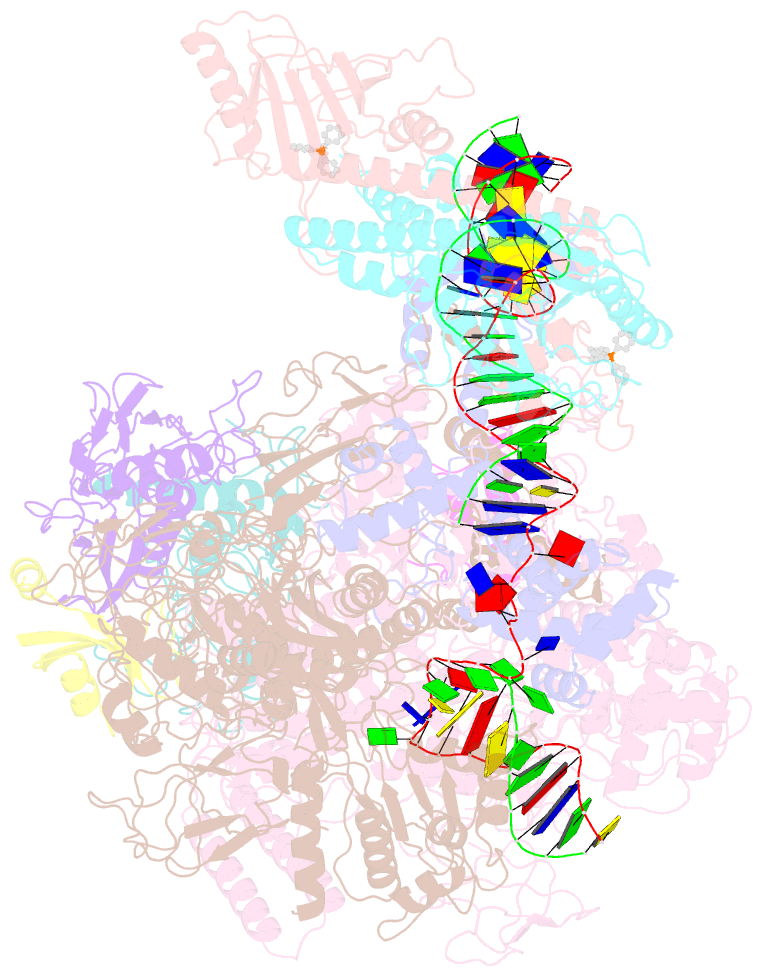
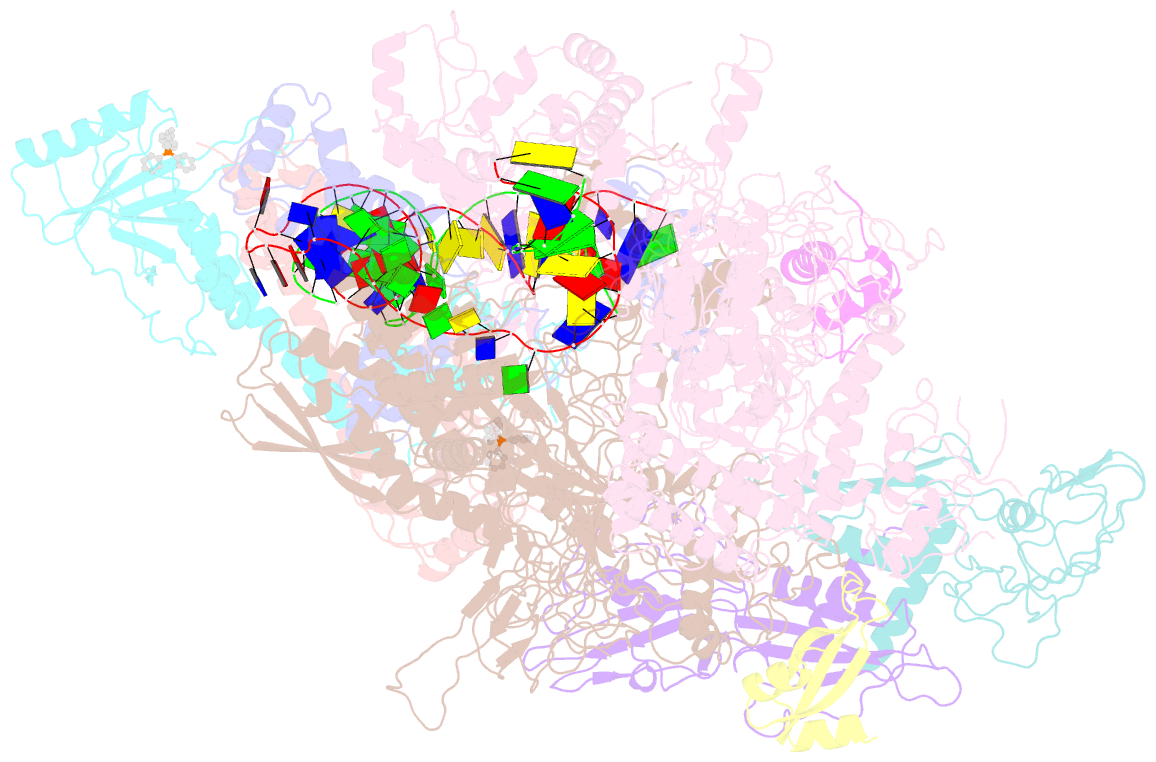
Summary information and primary citation
- PDB-id
- 7ckq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.4 Å)
- Summary
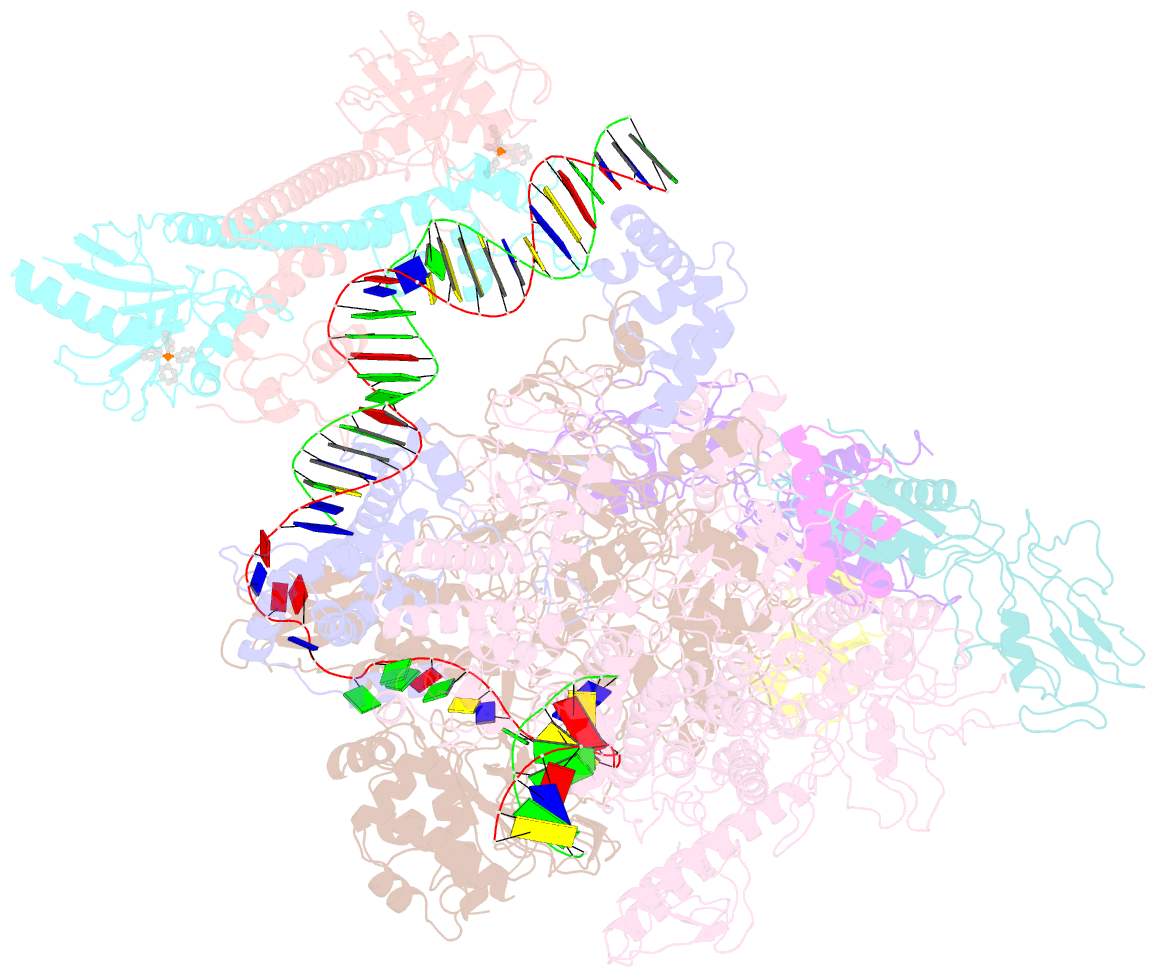
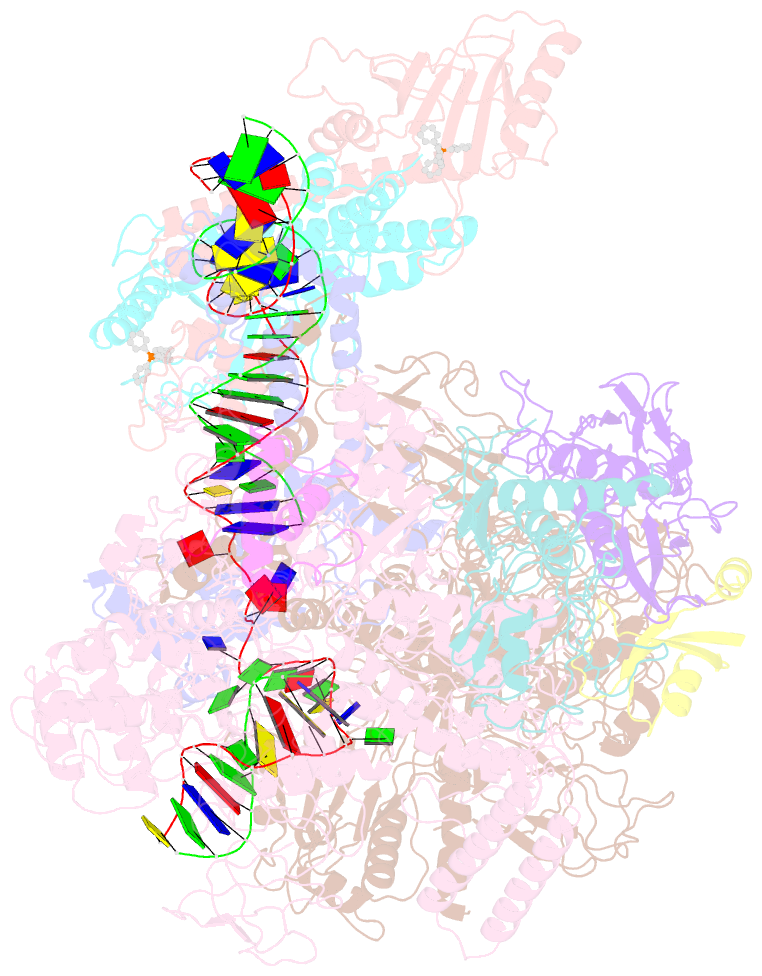
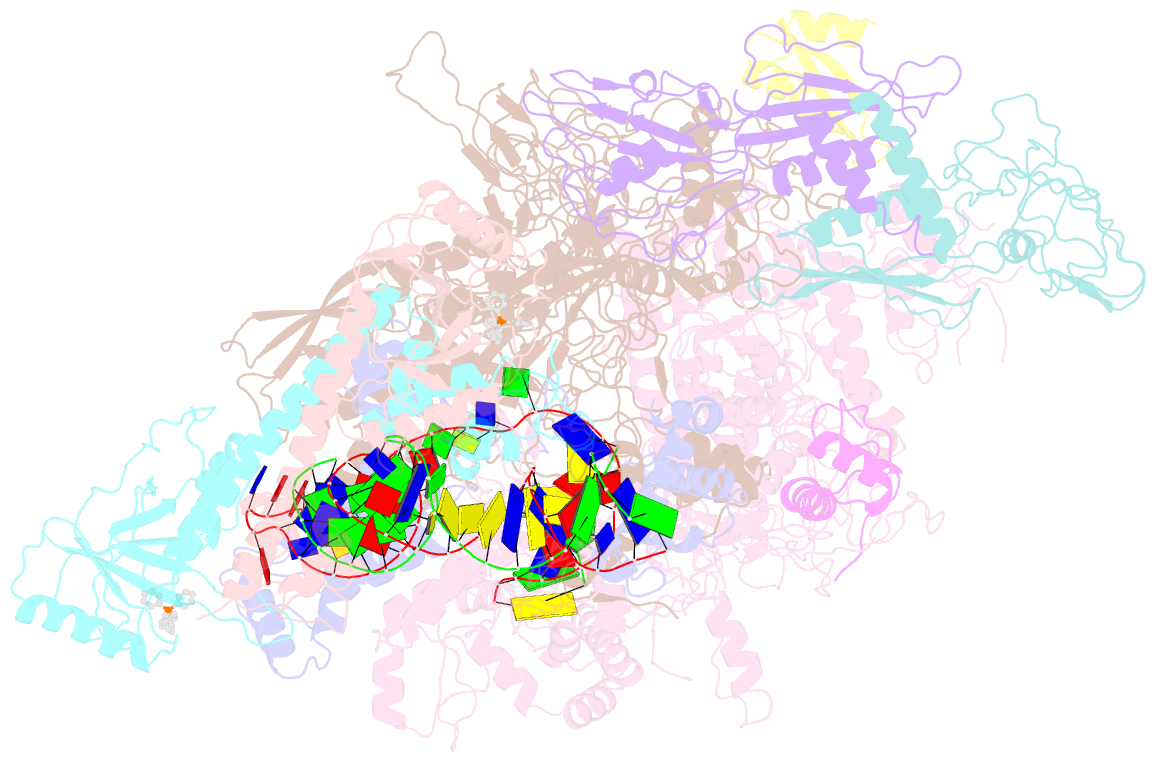
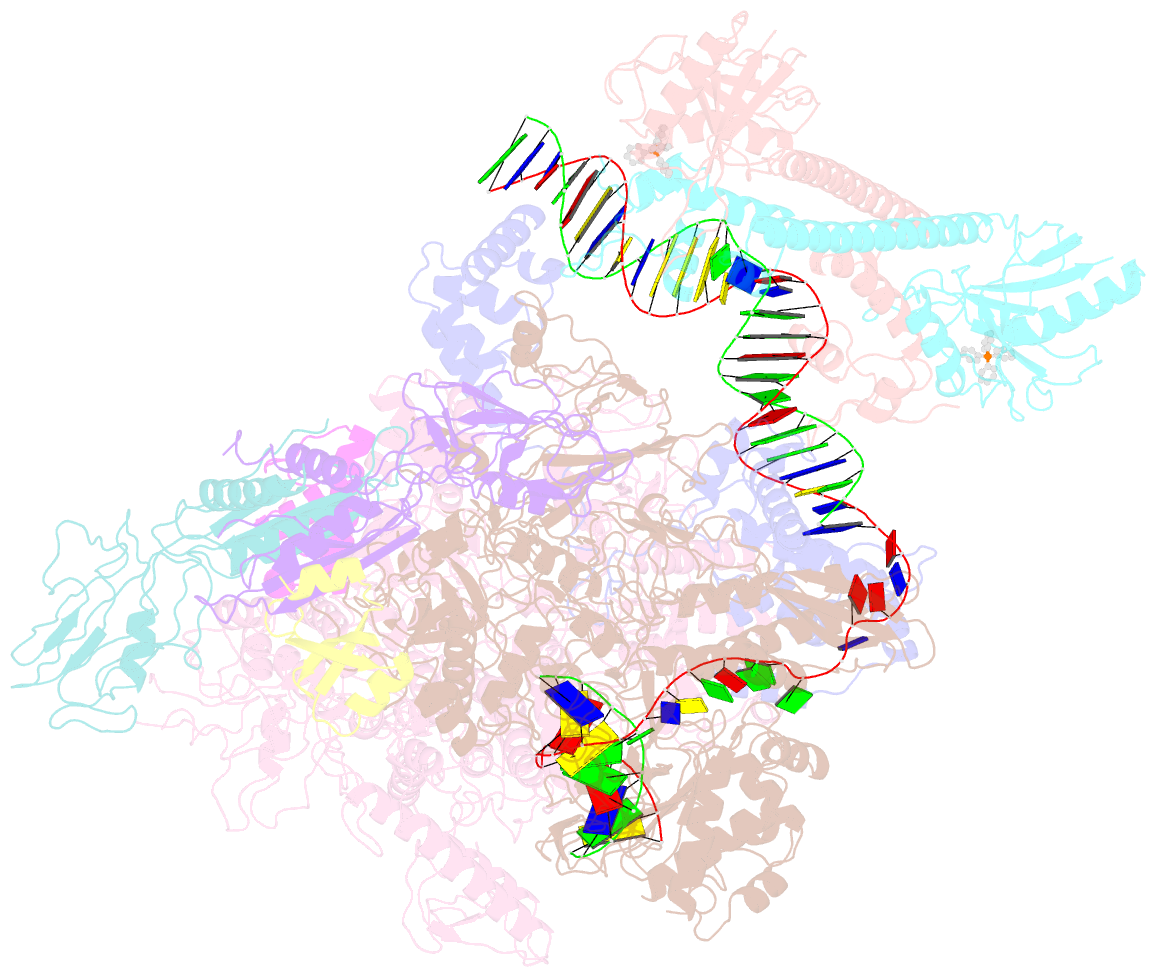
- The cryo-EM structure of b. subtilis bmrr transcription activation complex
- Reference
- Fang C, Li L, Zhao Y, Wu X, Philips SJ, You L, Zhong M, Shi X, O'Halloran TV, Li Q, Zhang Y (2020): "The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription." Nat Commun, 11, 6284. doi: 10.1038/s41467-020-20134-y.
- Abstract
- The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure of a B. subtilis transcription activation complex comprising B. subtilis six-subunit (2αββ'ωε) RNA Polymerase (RNAP) core enzyme, σA, a promoter DNA, and the ligand-bound B. subtilis BmrR, a prototype of MerR-family TFs. The structure reveals that RNAP and BmrR recognize the upstream promoter DNA from opposite faces and induce four significant kinks from the -35 element to the -10 element of the promoter DNA in a cooperative manner, which restores otherwise inactive promoter activity by shortening the length of promoter non-optimal -35/-10 spacer. Our structure supports a DNA-distortion and RNAP-non-contact paradigm of transcriptional activation by MerR TFs.