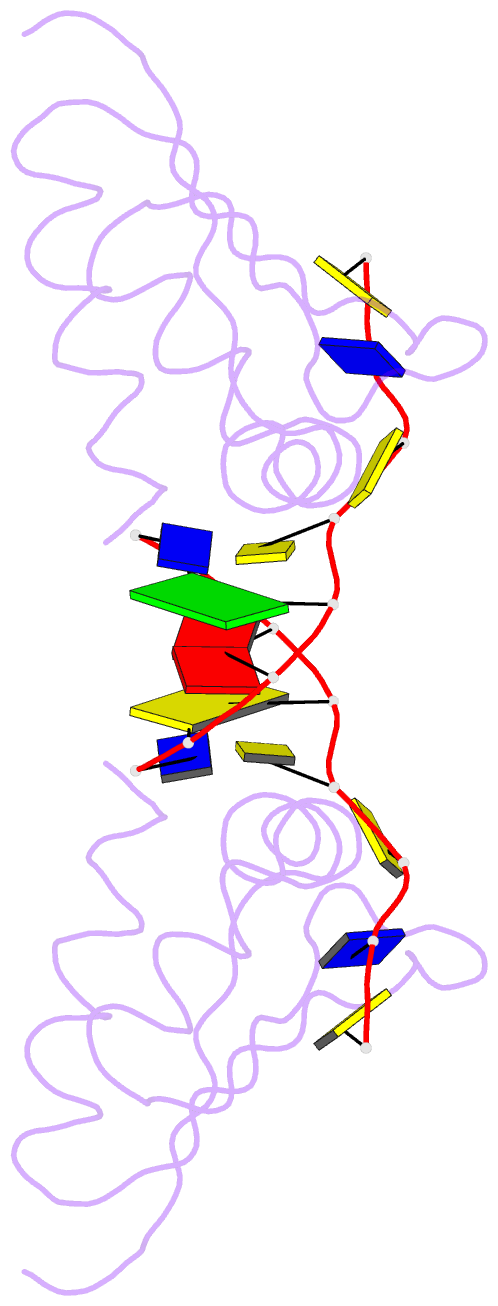
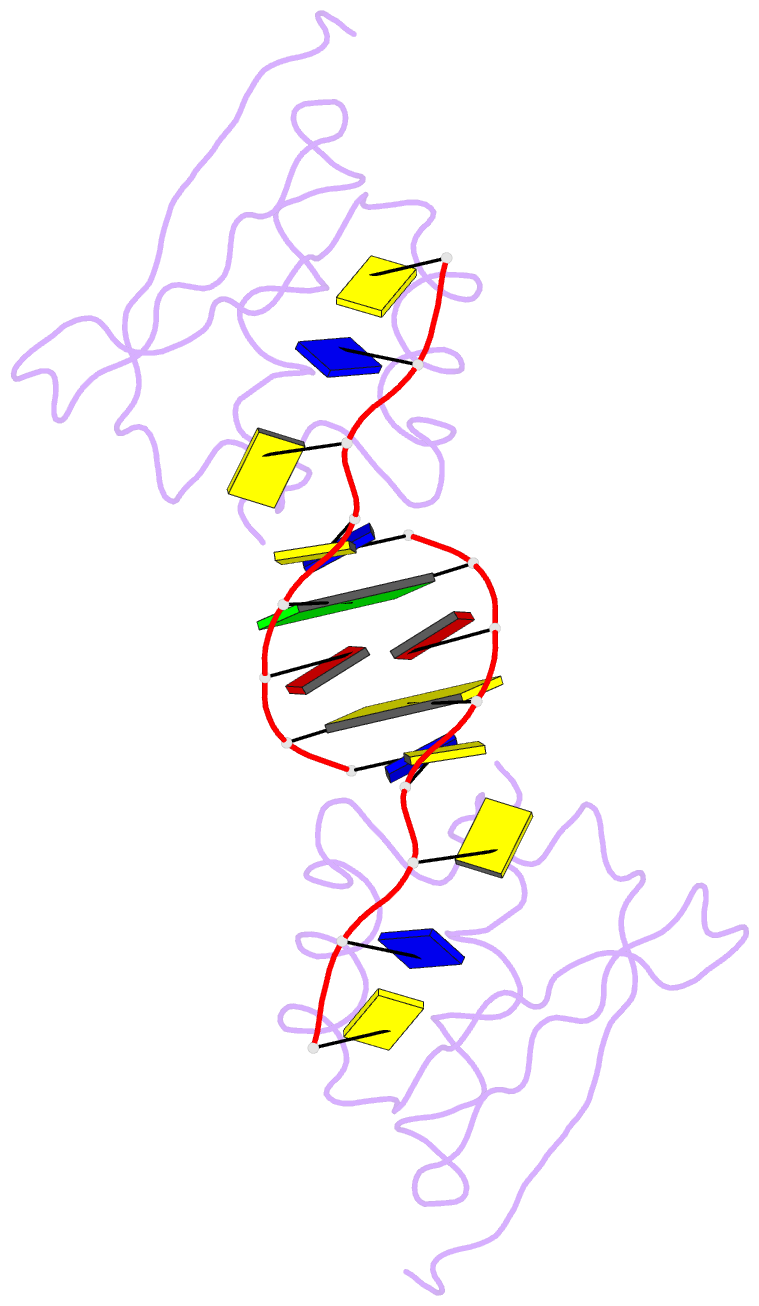
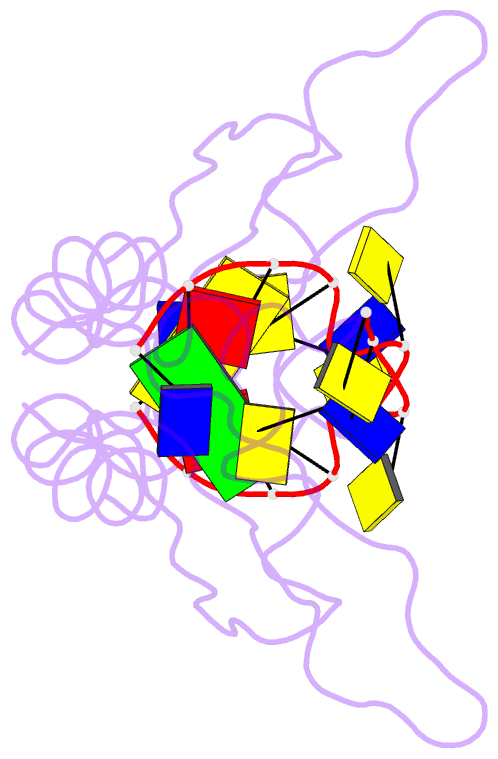
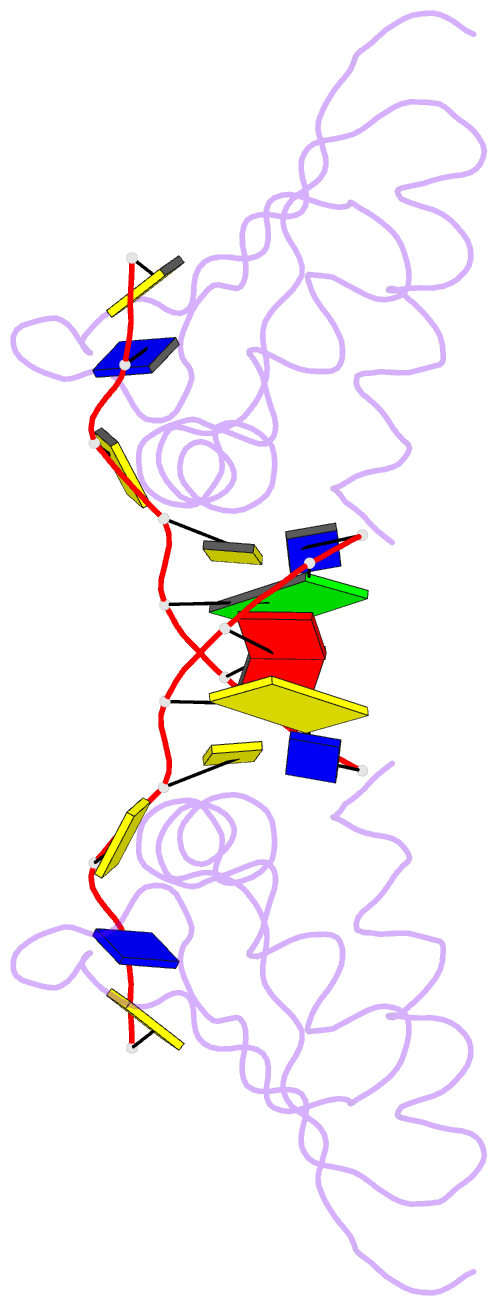
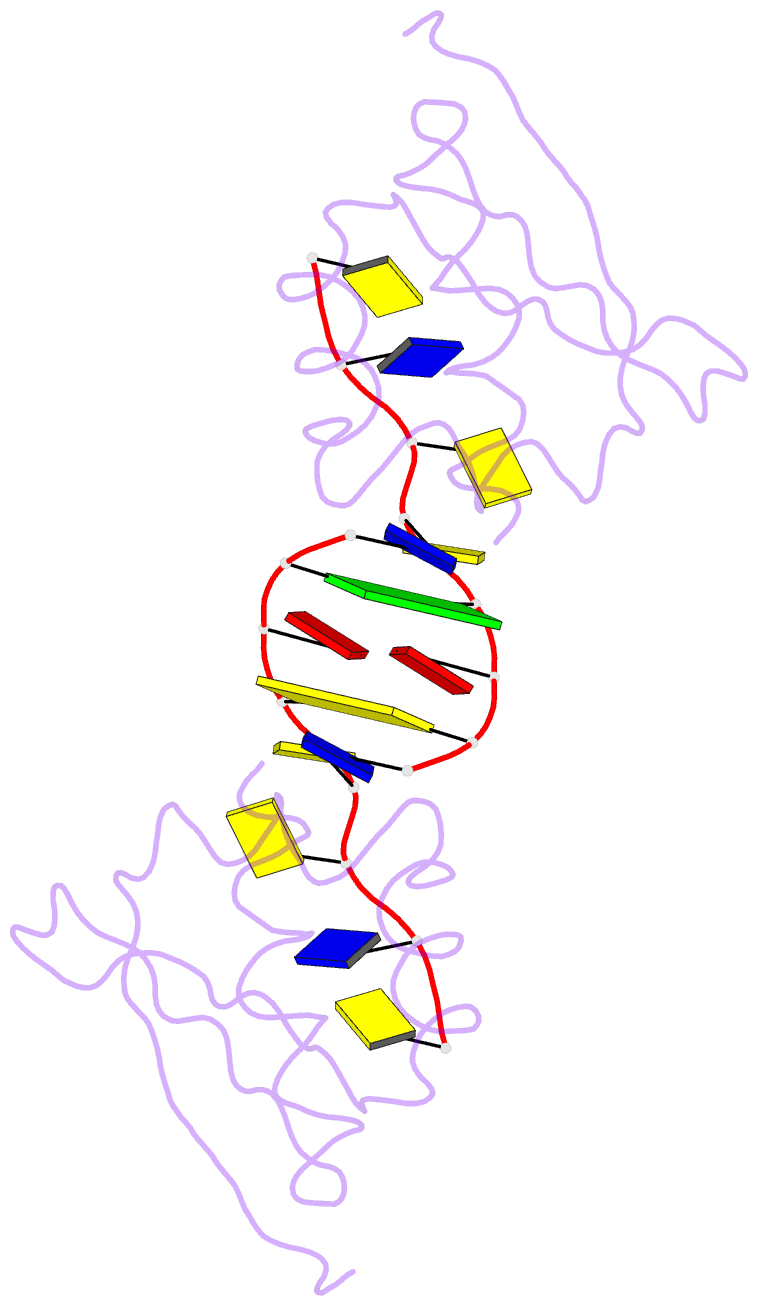
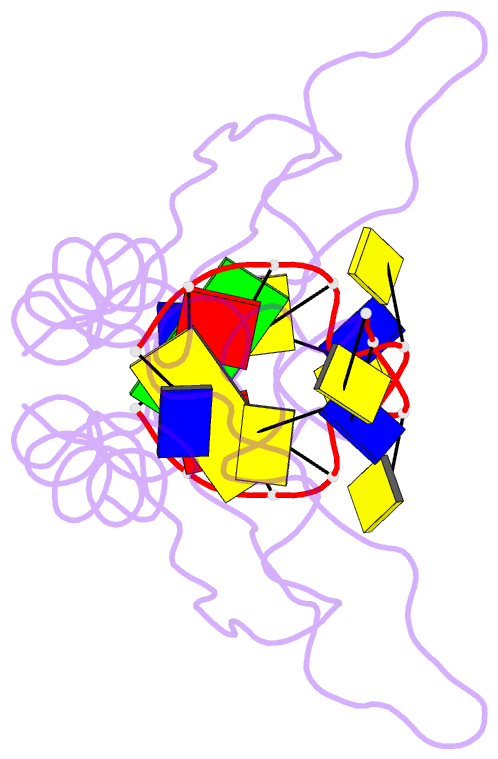
Summary information and primary citation
- PDB-id
- 7cre; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.0 Å)
- Summary
- Hnrnpk kh3 domain in complex with a ssDNA fragment from the sirloin element
- Reference
- Yao J, Tu Y, Shen C, Zhou Q, Xiao H, Jia D, Sun Q (2021): "Nuclear import receptors and hnRNPK mediates nuclear import and stress granule localization of SIRLOIN." Cell.Mol.Life Sci., 78, 7617-7633. doi: 10.1007/s00018-021-03992-7.
- Abstract
- The majority of lncRNAs and a small fraction of mRNAs localize in the cell nucleus to exert their functions. A SIRLOIN RNA motif was previously reported to drive its nuclear localization by the RNA-binding protein hnRNPK. However, the underlying mechanism remains unclear. Here, we report crystal structures of hnRNPK in complex with SIRLOIN, and with the nuclear import receptor (NIR) Impα1, respectively. The protein hnRNPK bound to SIRLOIN with multiple weak interactions, and interacted Impα1 using an independent high-affinity site. Forming a complex with hnRNPK and Impα1 was essential for the nuclear import and stress granule localization of SIRLOIN in semi-permeabilized cells. Nuclear import of SIRLOIN enhanced with increasing NIR concentrations, but its stress granule localization peaked at a low NIR concentration. Collectively, we propose a mechanism of SIRLOIN localization, in which NIRs functioned as drivers/regulators, and hnRNPK as an adaptor.