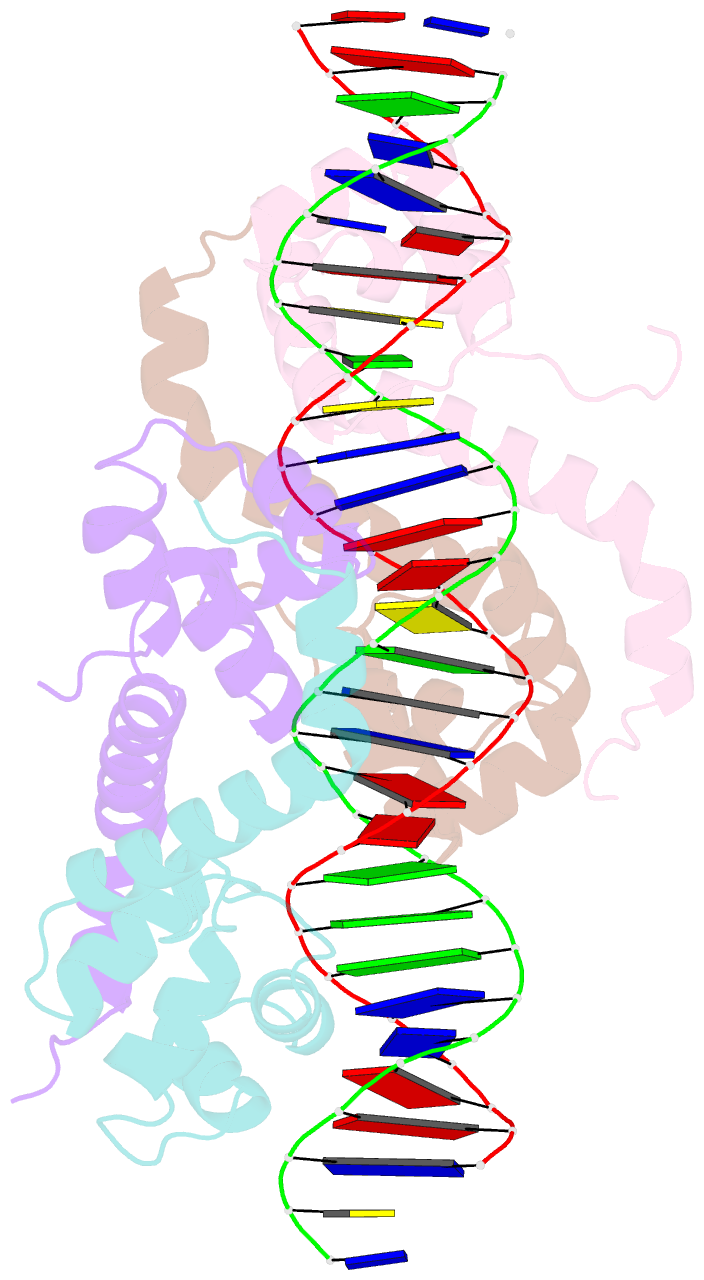
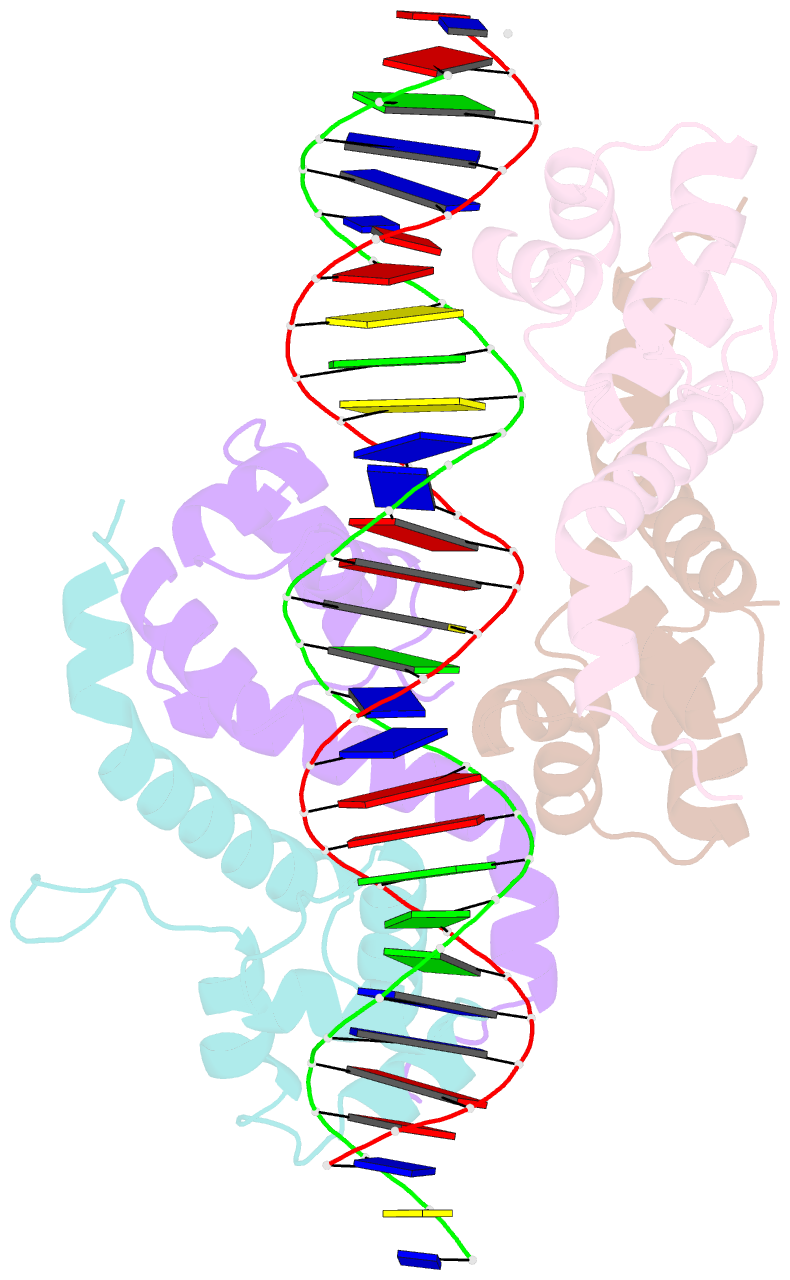
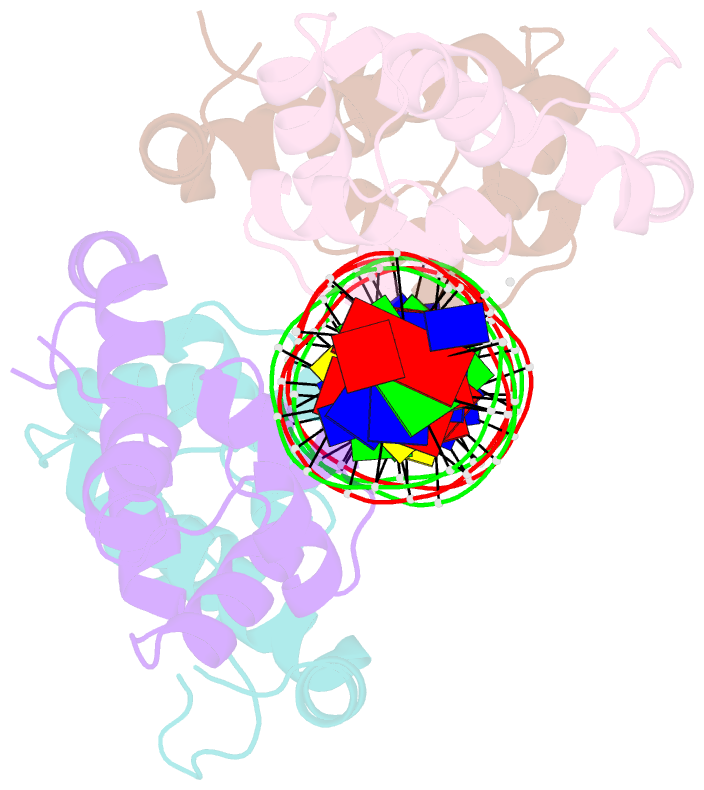
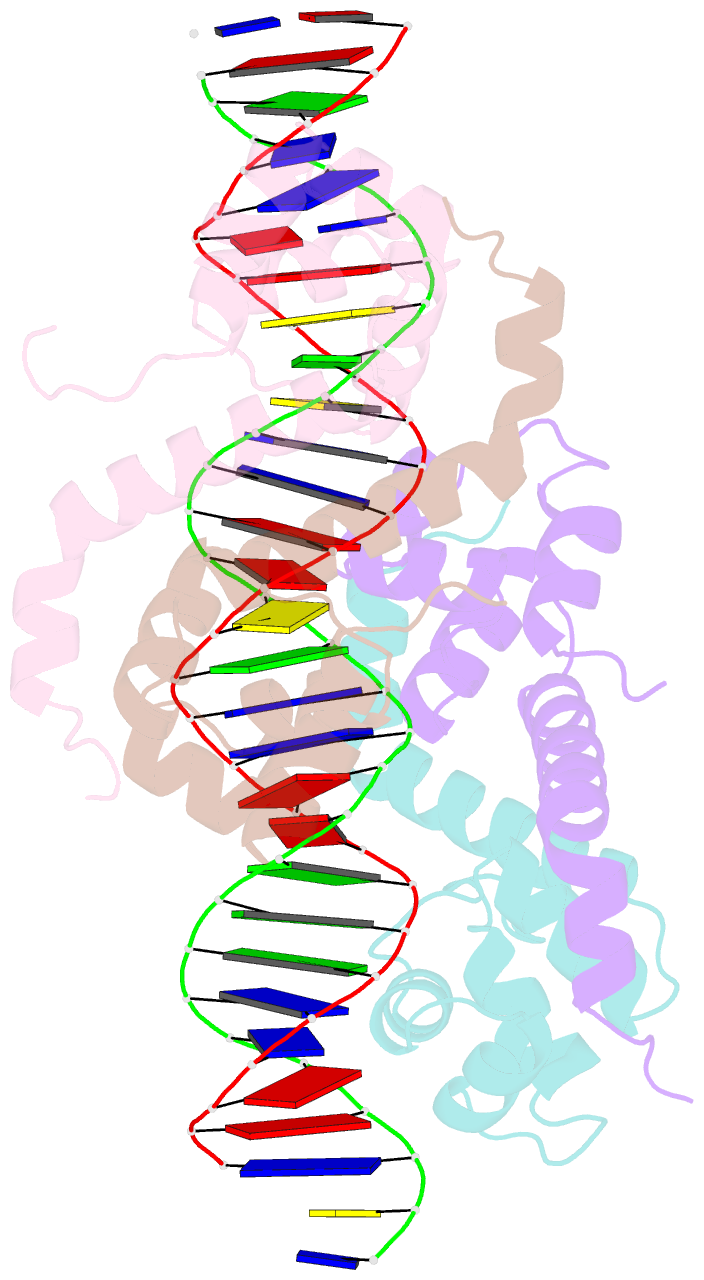
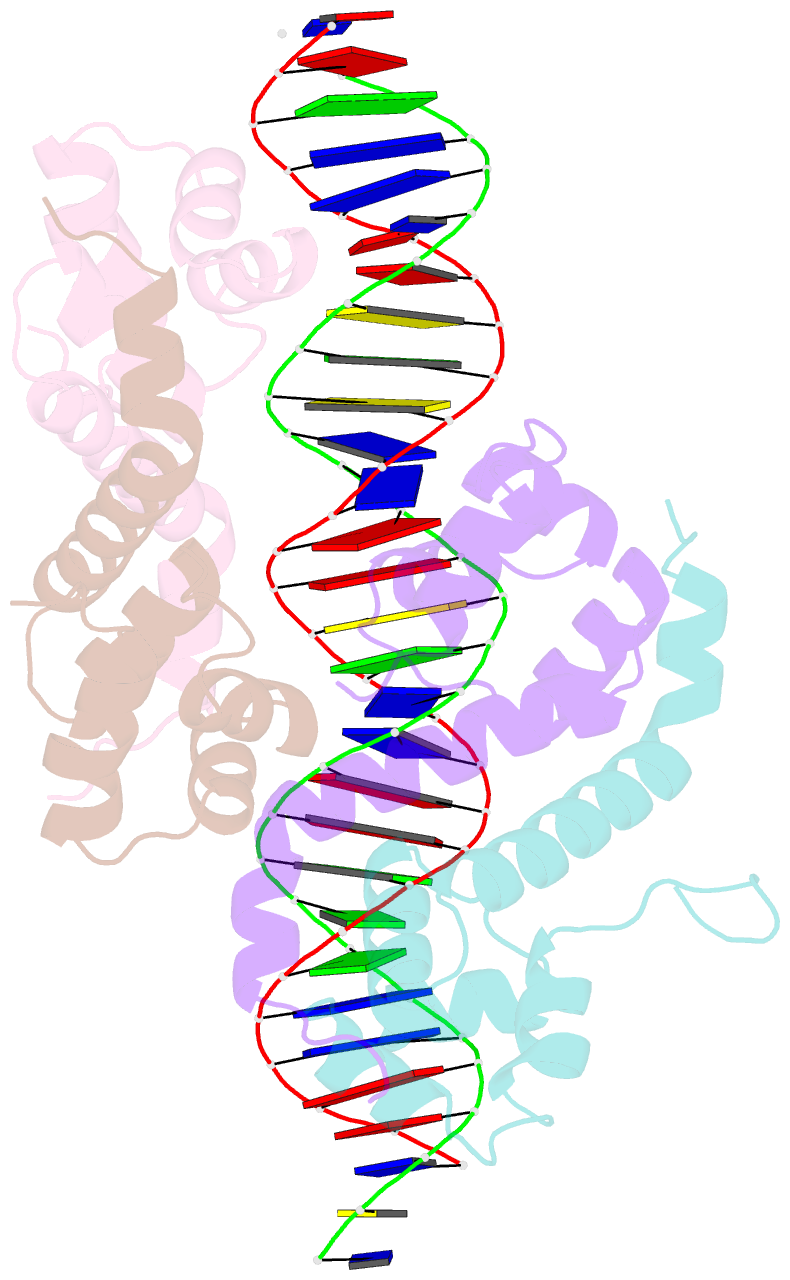
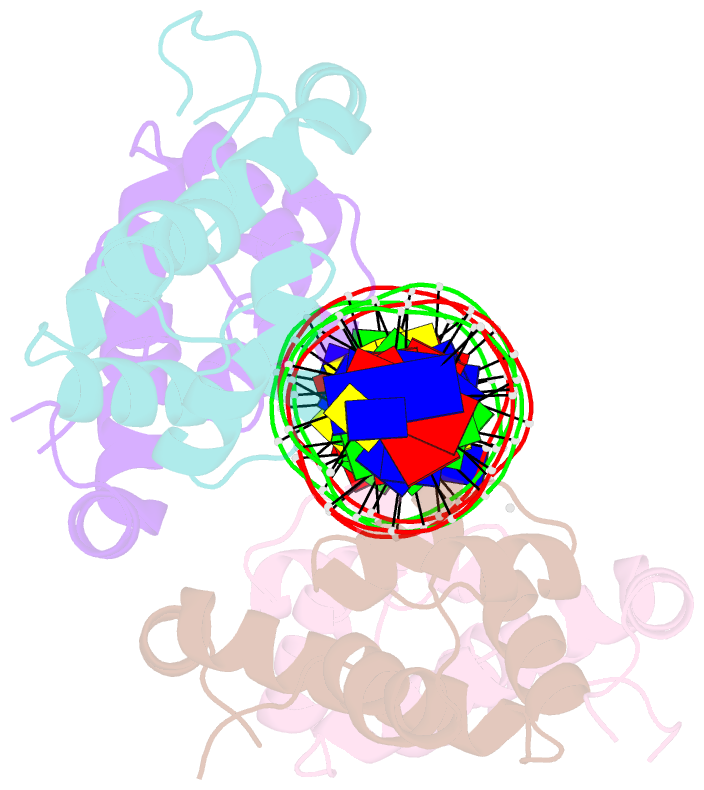
Summary information and primary citation
- PDB-id
- 7csy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- antitoxin-DNA
- Method
- X-ray (2.29 Å)
- Summary
- Pseudomonas aeruginosa antitoxin higa with higba promoter
- Reference
- Song Y, Luo G, Zhu Y, Li T, Li C, He L, Zhao N, Zhao C, Yang J, Huang Q, Mu X, Tang X, Kang M, Wu S, He Y, Bao R (2021): "Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs." Environ.Microbiol., 23, 1541-1558. doi: 10.1111/1462-2920.15365.
- Abstract
- Type II toxin-antitoxin (TA) systems modulate many essential cellular processes in prokaryotic organisms. Recent studies indicate certain type II antitoxins also transcriptionally regulate other genes, besides neutralizing toxin activity. Herein, we investigated the diverse transcriptional repression properties of type II TA antitoxin PaHigA from Pseudomonas aeruginosa. Biochemical and functional analyses showed that PaHigA recognized variable pseudopalindromic DNA sequences and repressed expression of multiple genes. Furthermore, we presented high resolution structures of apo-PaHigA, PaHigA-PhigBA and PaHigA-Ppa2440 complex, describing how the rearrangements of the HTH domain accounted for the different DNA-binding patterns among HigA homologues. Moreover, we demonstrated that the N-terminal loop motion of PaHigA was associated with its apo and DNA-bound states, reflecting a switch mechanism regulating HigA antitoxin function. Collectively, this work extends our understanding of how the PaHigB/HigA system regulates multiple metabolic pathways to balance the growth and stress response in P. aeruginosa and could guide further development of anti-TA oriented strategies for pathogen treatment.