Summary information and primary citation
- PDB-id
- 7csz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-DNA
- Method
- X-ray (1.8 Å)
- Summary
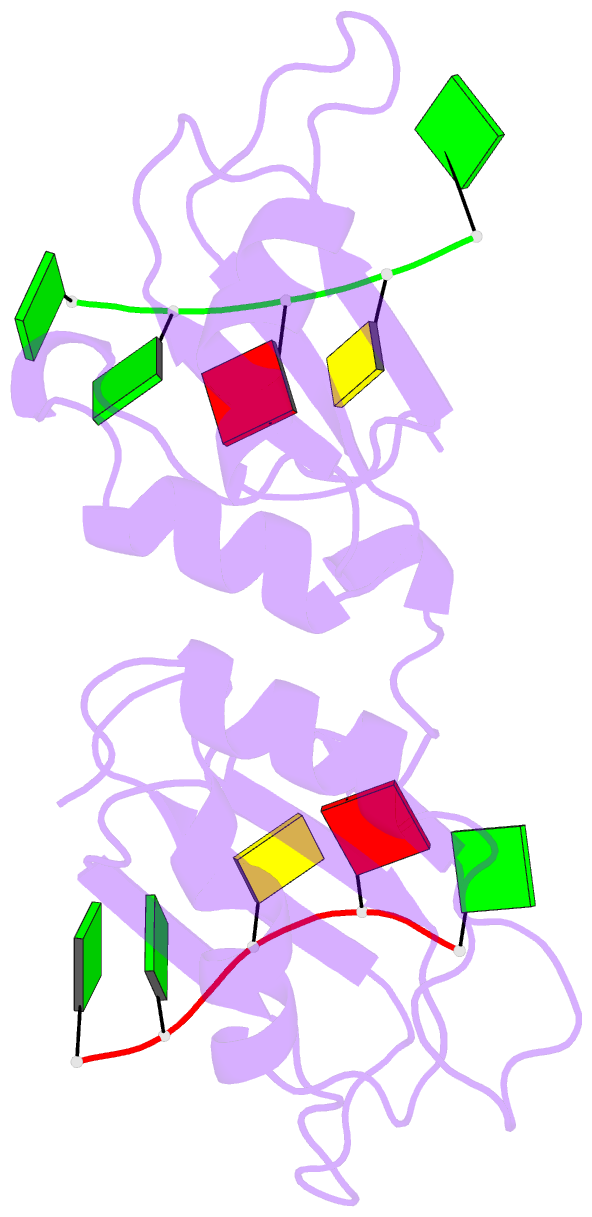
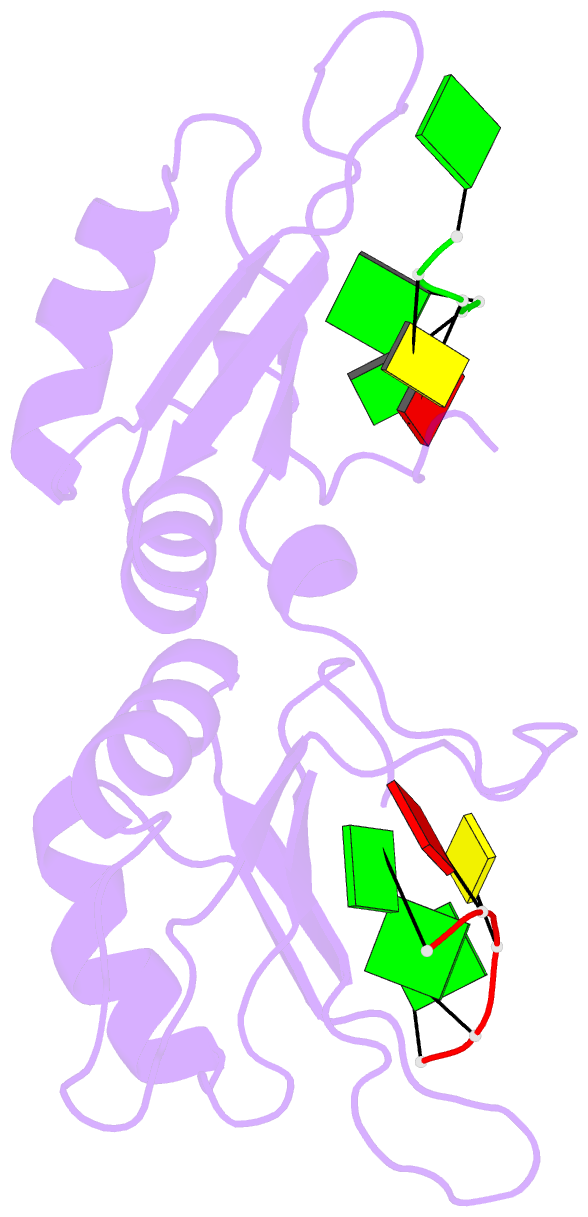
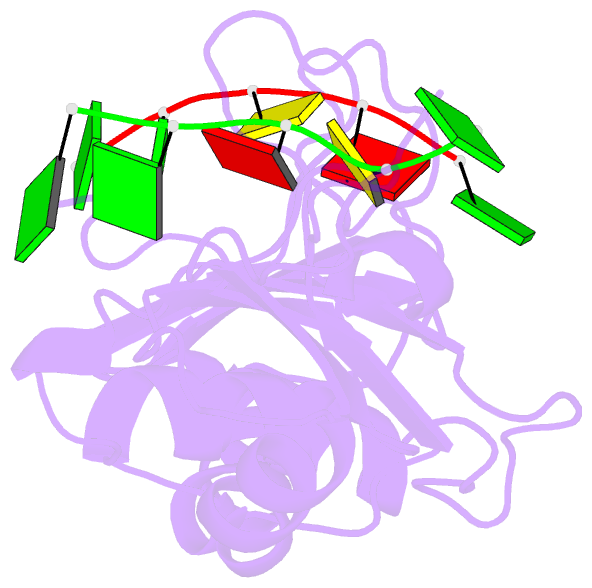
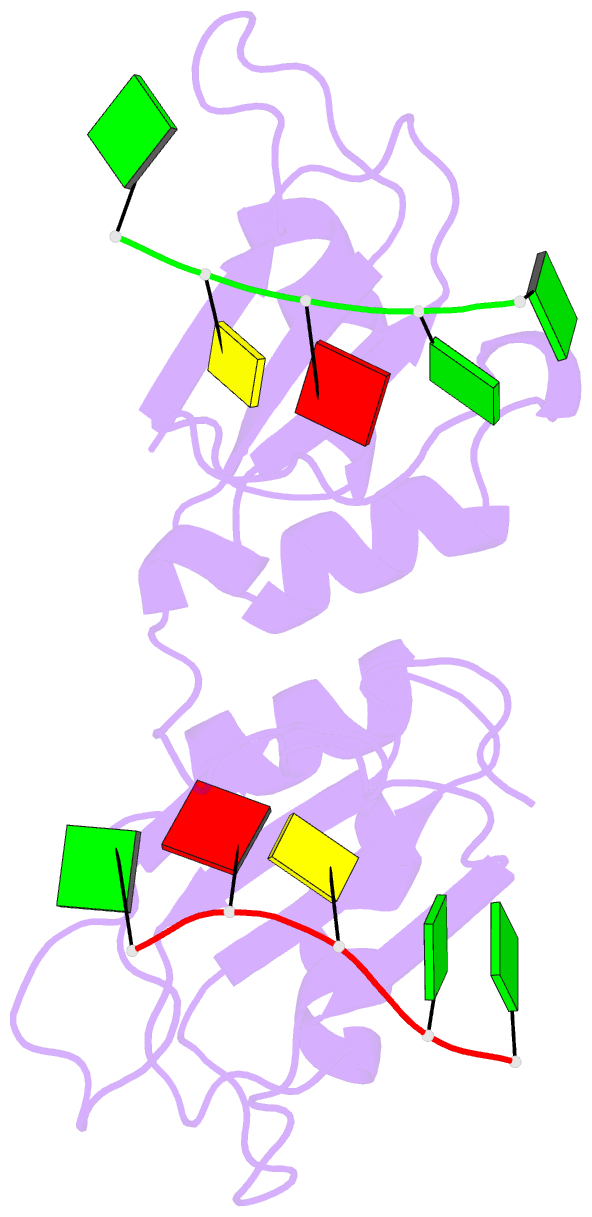
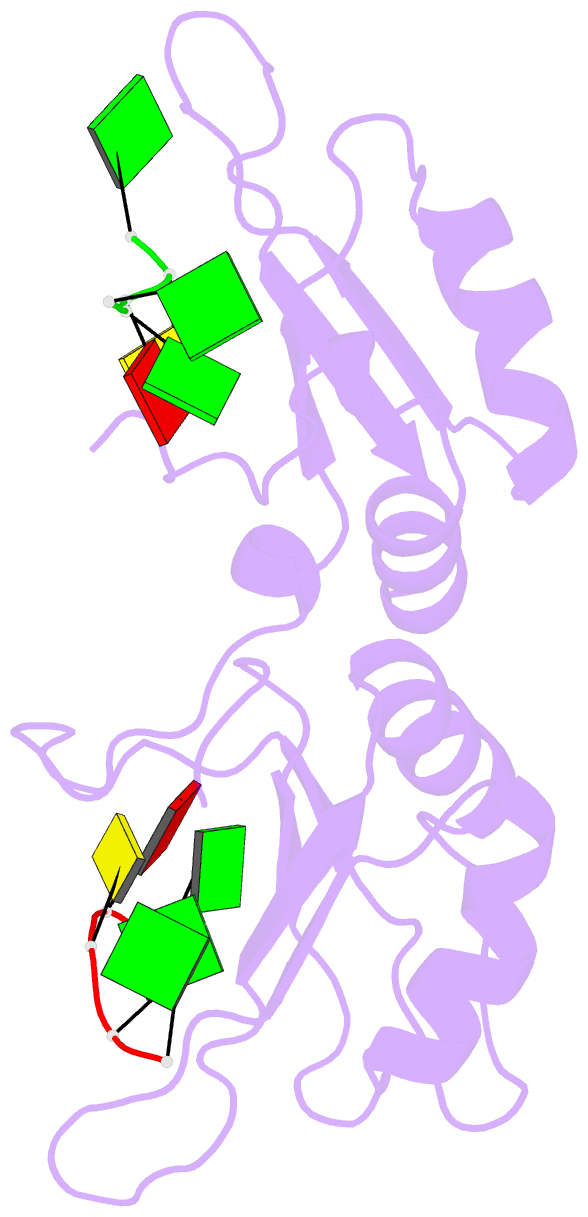
- Crystal structure of the n-terminal tandem rrm domains of rbm45 in complex with single-stranded DNA
- Reference
- Chen X, Yang Z, Wang W, Qian K, Liu M, Wang J, Wang M (2021): "Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45." Nucleic Acids Res., 49, 2946-2958. doi: 10.1093/nar/gkab075.
- Abstract
- RBM45 is an RNA-binding protein involved in neural development, whose aggregation is associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD). However, the mechanisms of RNA-binding and aggregation of RBM45 remain unelucidated. Here, we report the crystal structure of the N-terminal tandem RRM domains of human RBM45 in complex with single-stranded DNA (ssDNA). Our structural and biochemical results revealed that both the RRM1 and RRM2 of RBM45 recognized the GAC sequence of RNA/ssDNA. Two aromatic residues and an arginine residue in each RRM were critical for RNA-binding, and the interdomain linker was also involved in RNA-binding. Two RRMs formed a pair of antiparallel RNA-binding sites, indicating that the N-terminal tandem RRM domains of RBM45 bound separate GAC motifs in one RNA strand or GAC motifs in different RNA strands. Our findings will be helpful in the identification of physiologic targets of RBM45 and provide evidence for understanding the physiologic and pathologic functions of RBM45.