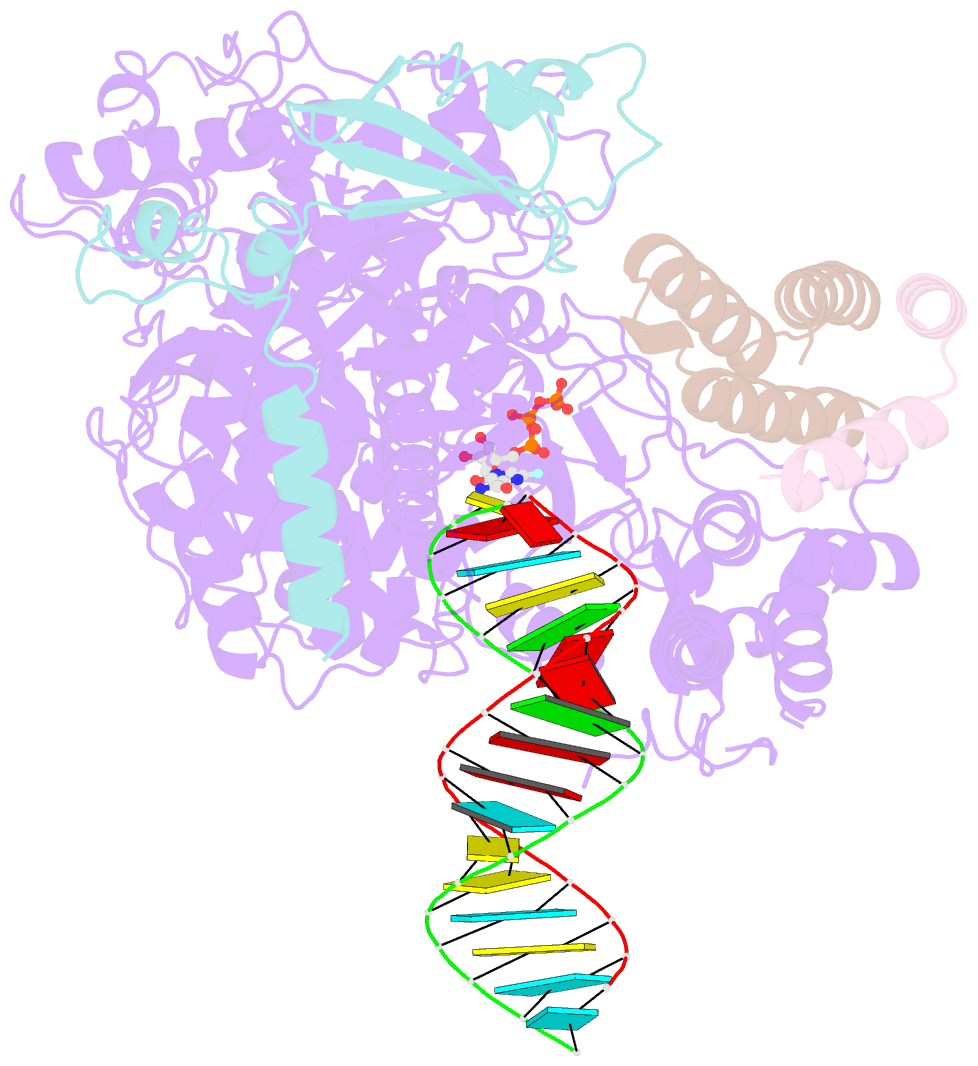
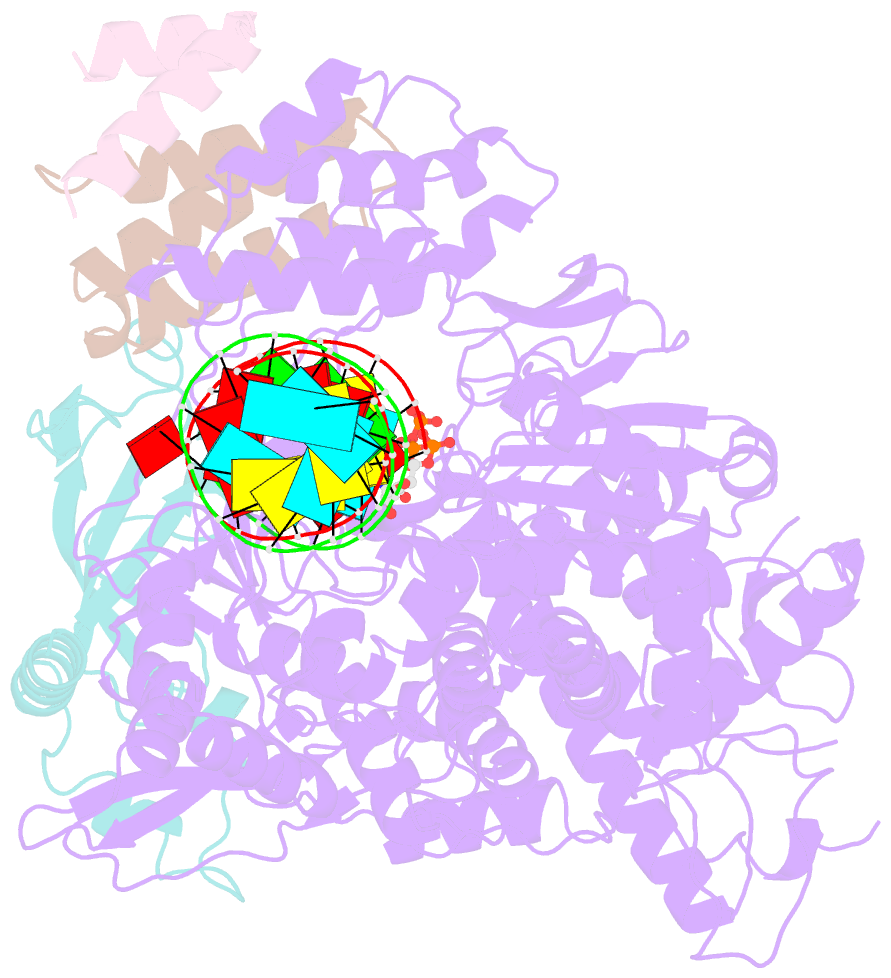
Summary information and primary citation
- PDB-id
- 7ctt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.2 Å)
- Summary
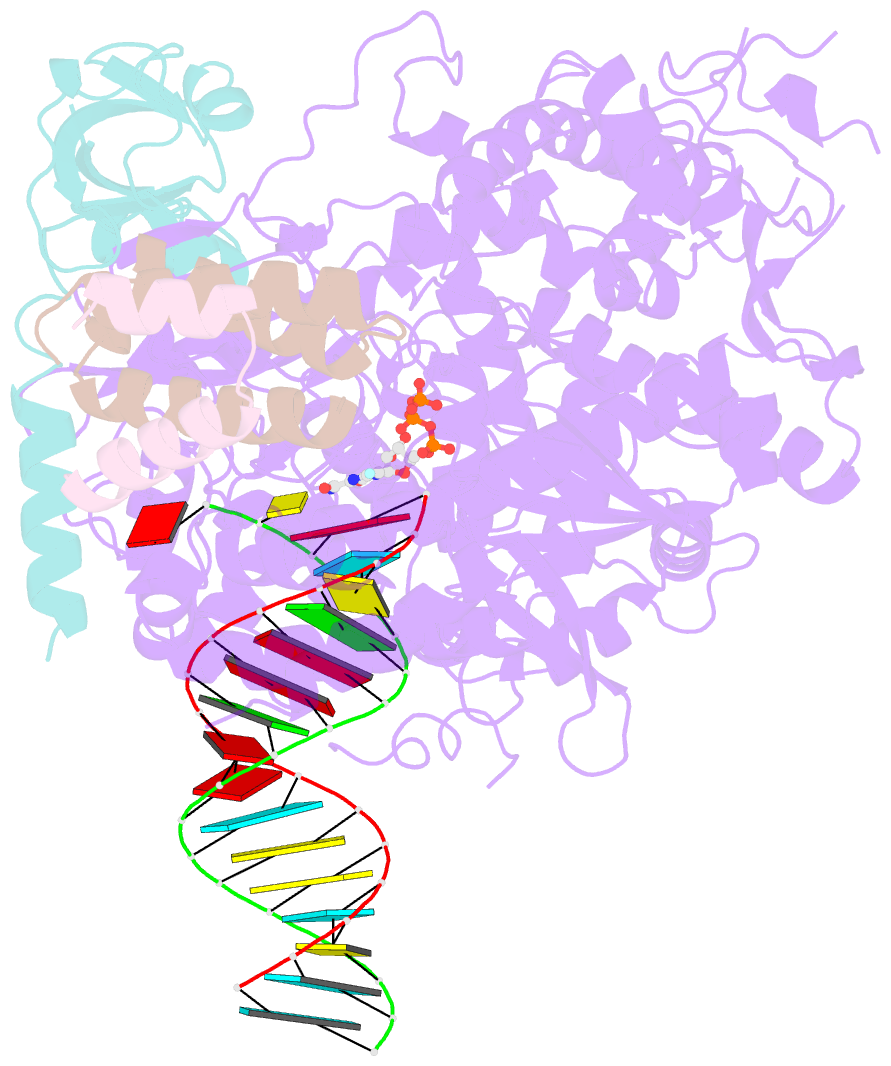
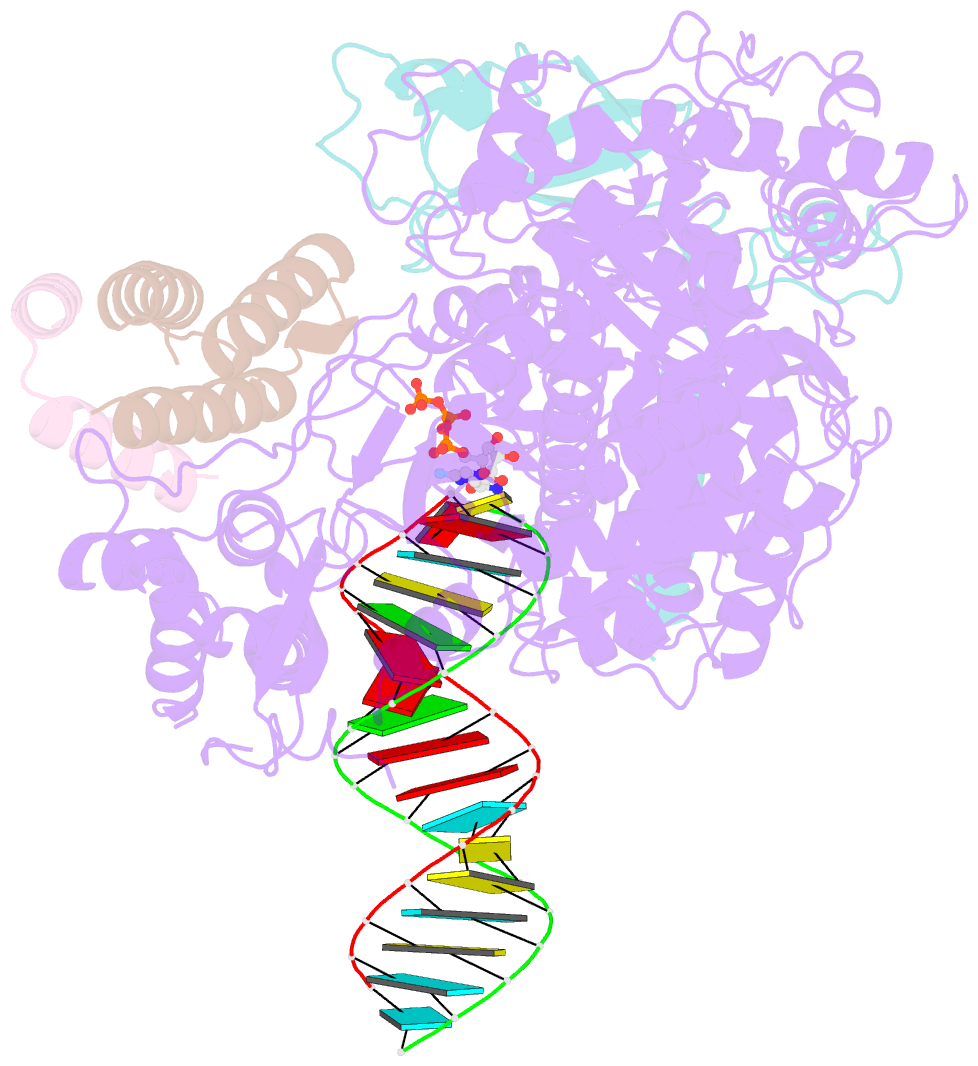
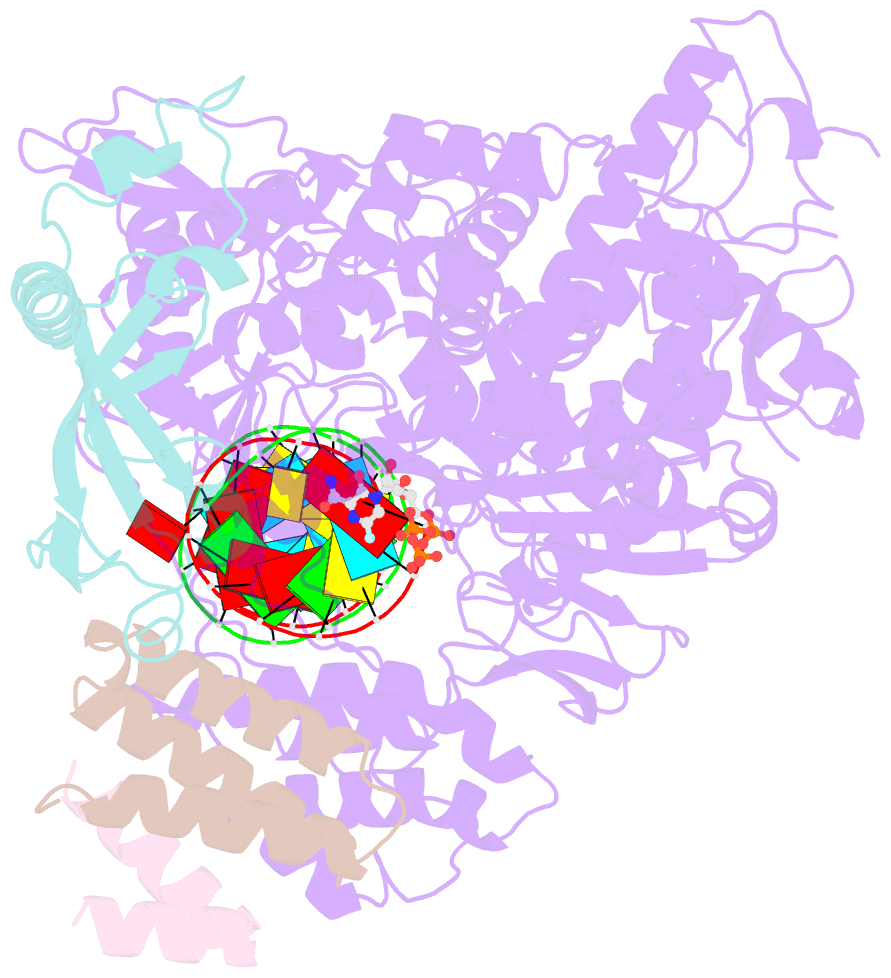
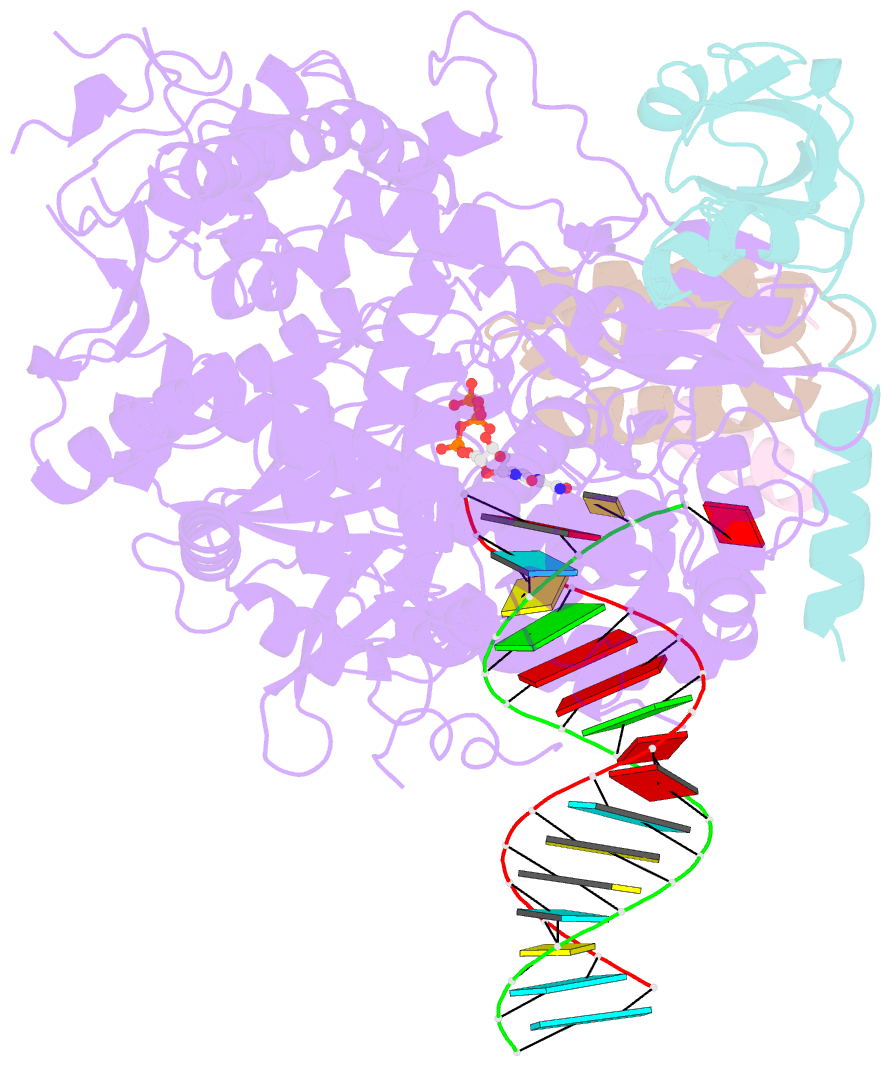
- cryo-EM structure of favipiravir bound to replicating polymerase complex of sars-cov-2 in the pre-catalytic state.
- Reference
- Peng Q, Peng R, Yuan B, Wang M, Zhao J, Fu L, Qi J, Shi Y (2021): "Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir." Innovation (N Y), 2, 100080. doi: 10.1016/j.xinn.2021.100080.
- Abstract
- The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has developed into an unprecedented global pandemic. Nucleoside analogs, such as Remdesivir and Favipiravir, can serve as the first-line broad-spectrum antiviral drugs by targeting the viral polymerases. However, the underlying mechanisms for the antiviral efficacies of these drugs are far from well understood. Here, we reveal that Favipiravir, as a pyrazine derivative, could be incorporated into the viral RNA products by mimicking both adenine and guanine nucleotides. This drug thus inhibits viral replication mainly by inducing mutations in progeny RNAs, different from Remdesivir or other RNA-terminating nucleoside analogs that impair the elongation of RNA products. We further determined the cryo-EM structure of Favipiravir bound to the replicating polymerase complex of SARS-CoV-2 in the pre-catalytic state. This structure provides a missing snapshot for visualizing the catalysis dynamics of coronavirus polymerase, and reveals an unexpected base-pairing pattern between Favipiravir and pyrimidine residues that may explain its capacity for mimicking both adenine and guanine nucleotides. These findings shed light on the mechanism of coronavirus polymerase catalysis and provide a rational basis for developing antiviral drugs to combat the SARS-CoV-2 pandemic.