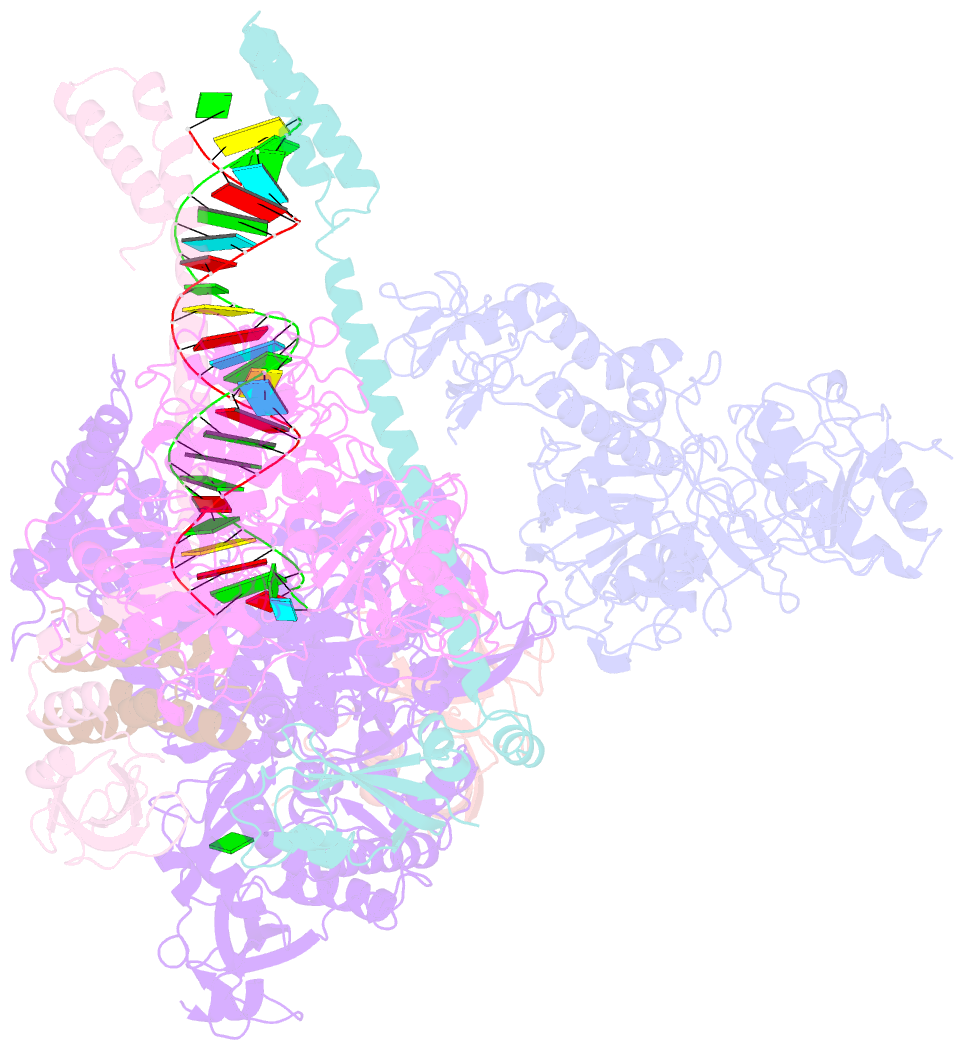
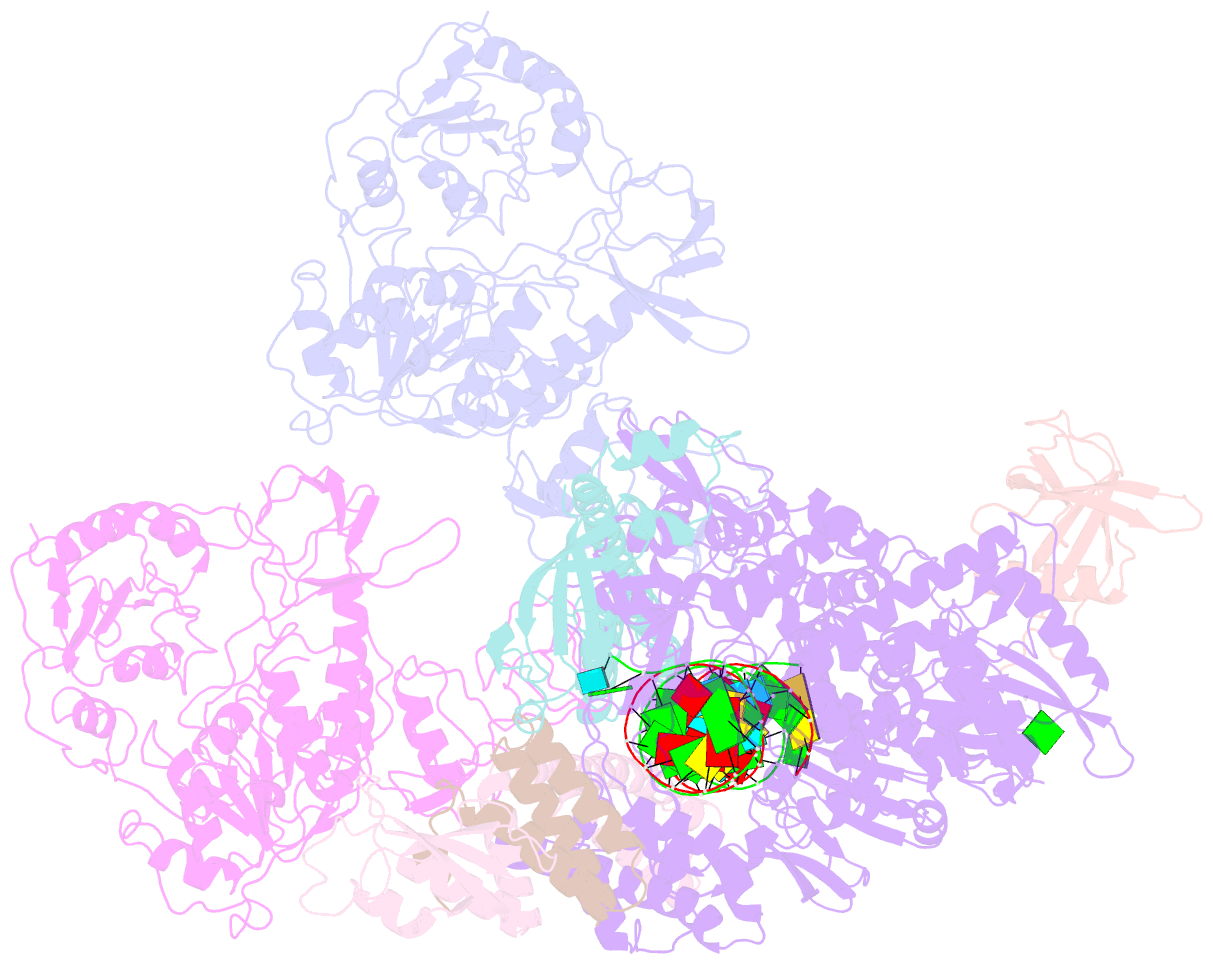
Summary information and primary citation
- PDB-id
- 7cyq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- cryo-EM (2.83 Å)
- Summary
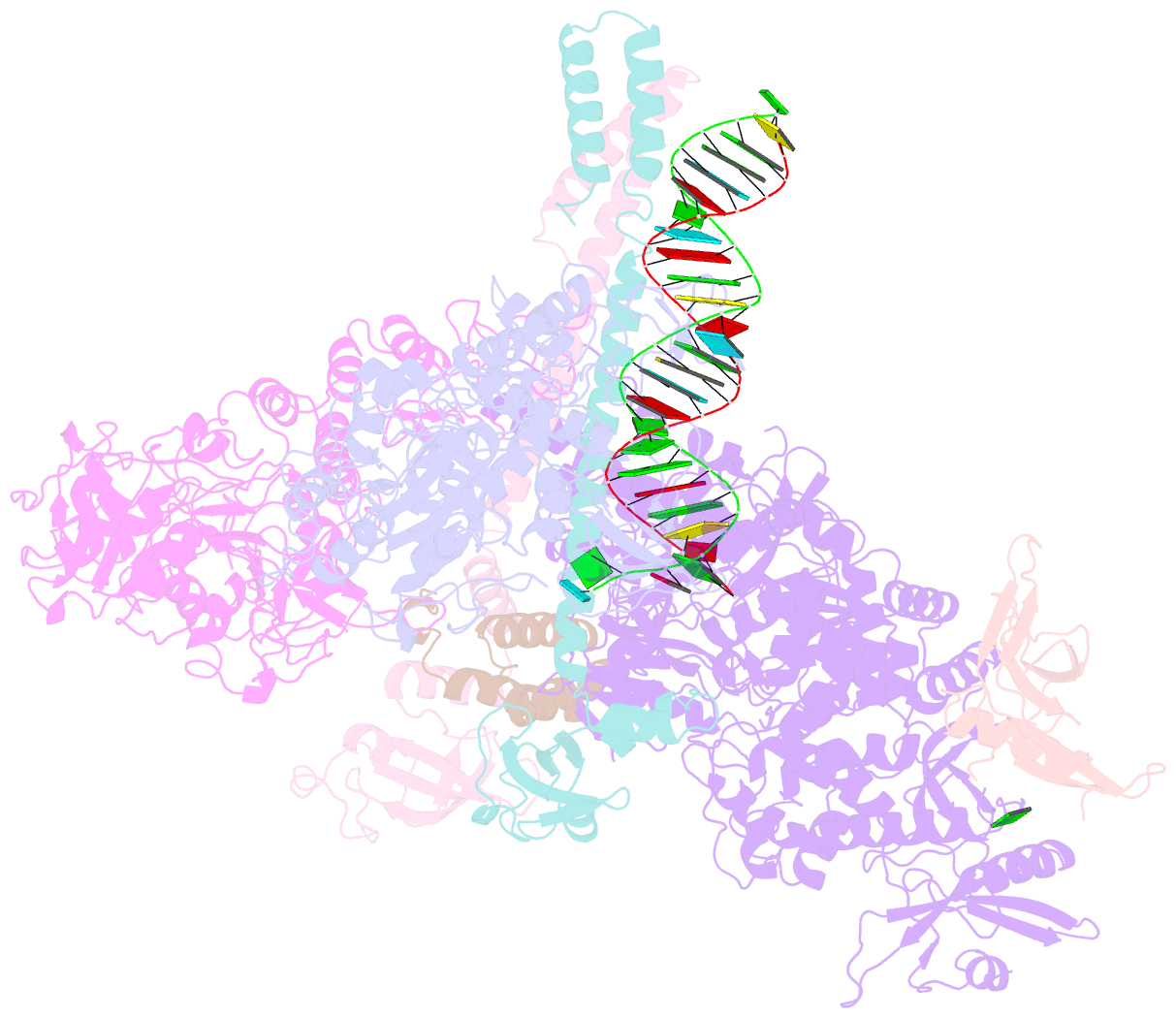
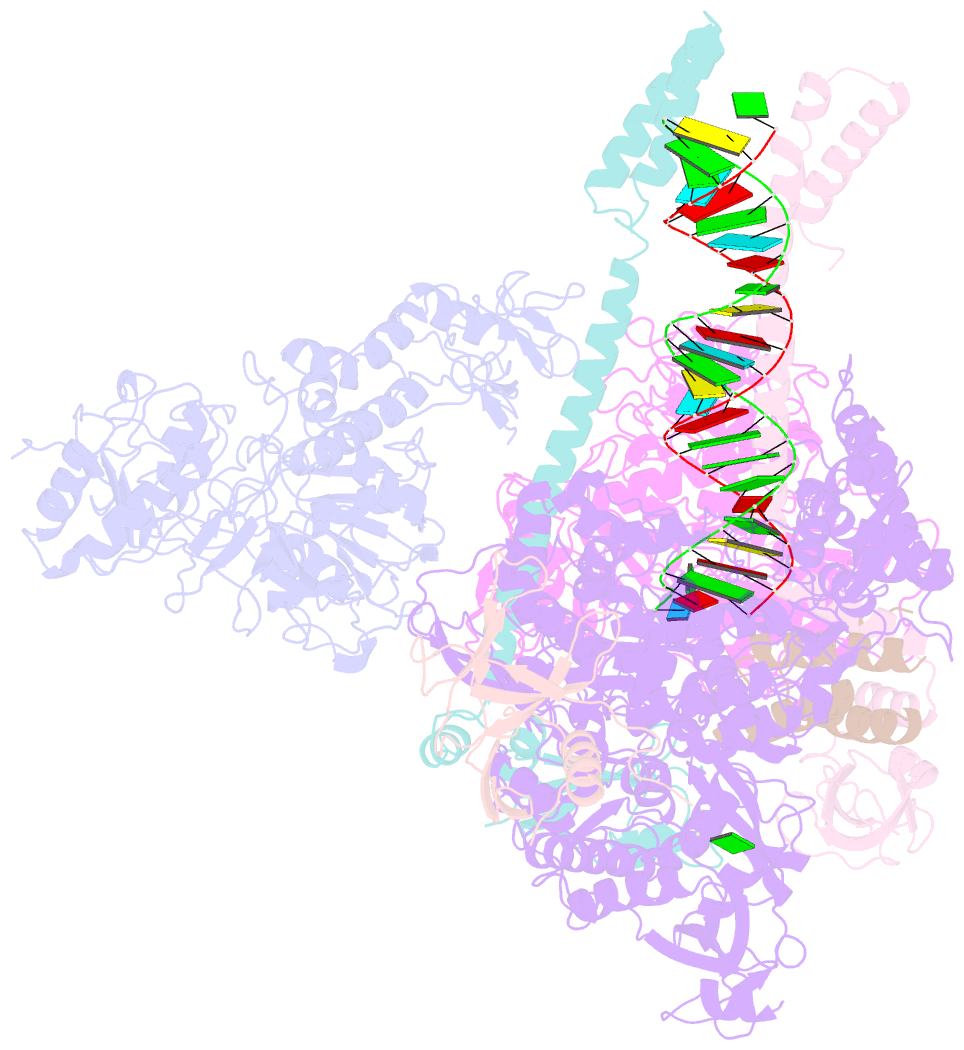
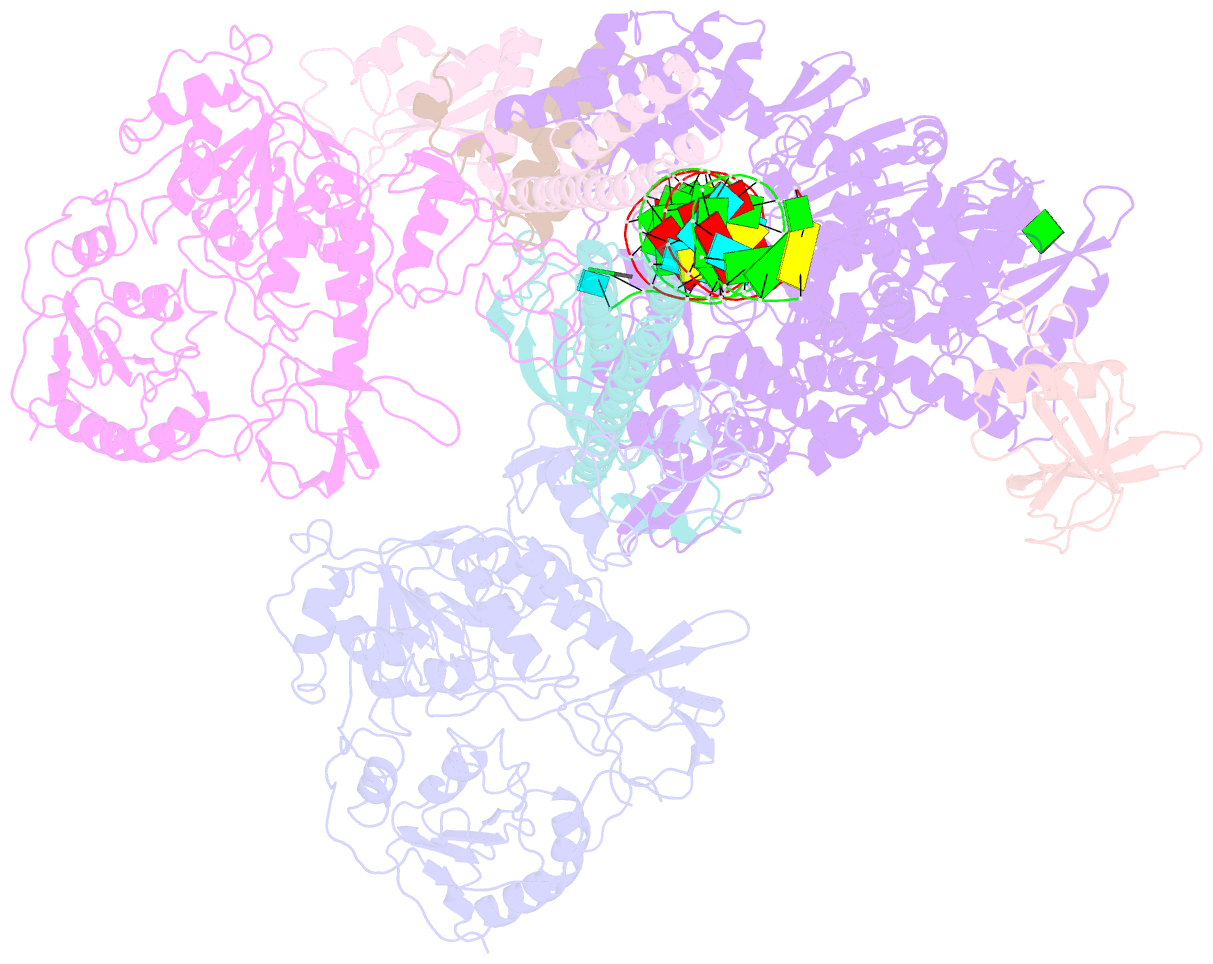
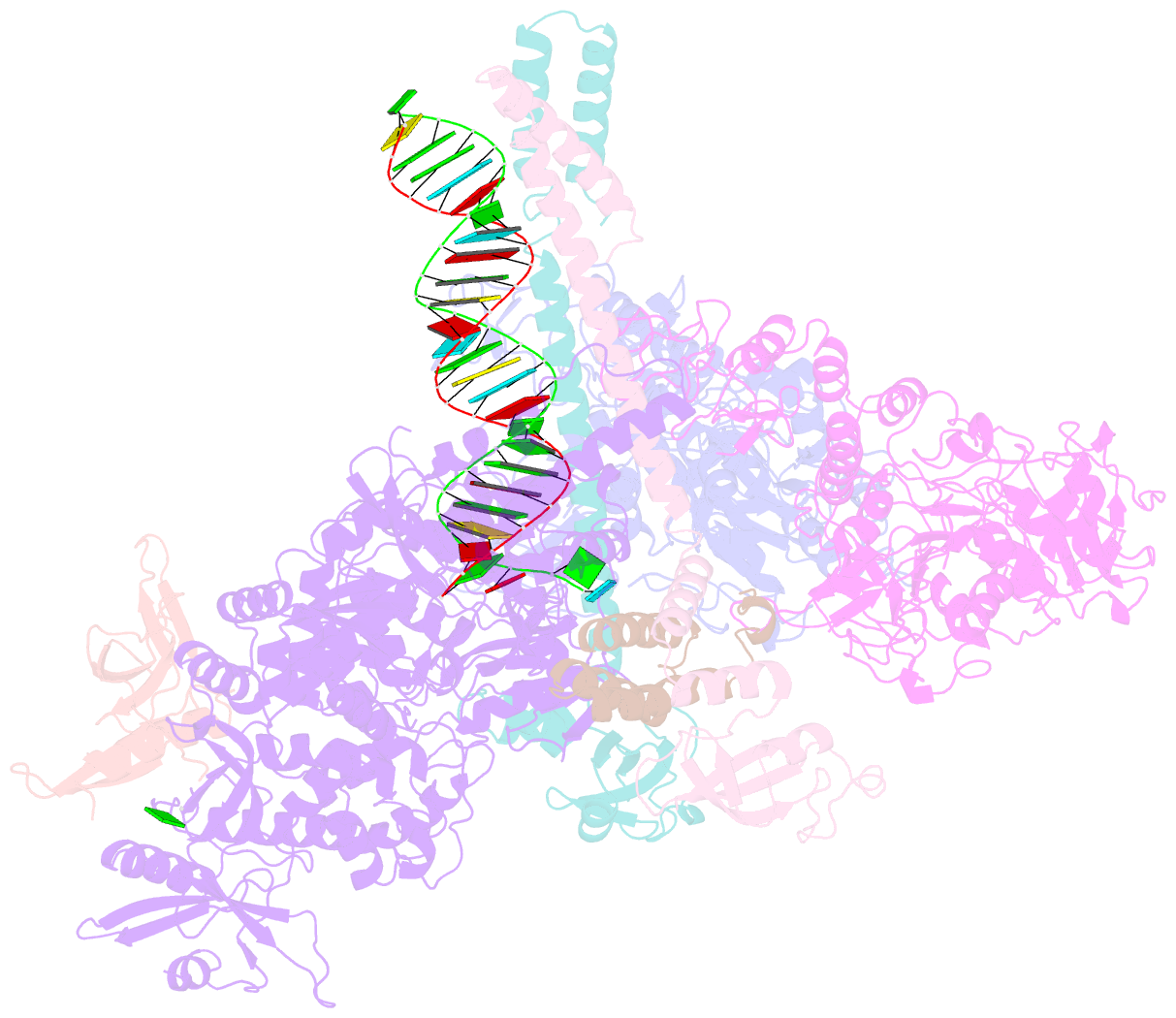
- cryo-EM structure of an extended sars-cov-2 replication and transcription complex reveals an intermediate state in cap synthesis
- Reference
- Yan L, Ge J, Zheng L, Zhang Y, Gao Y, Wang T, Huang Y, Yang Y, Gao S, Li M, Liu Z, Wang H, Li Y, Chen Y, Guddat LW, Wang Q, Rao Z, Lou Z (2021): "Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis." Cell, 184, 184-193.e10. doi: 10.1016/j.cell.2020.11.016.
- Abstract
- Transcription of SARS-CoV-2 mRNA requires sequential reactions facilitated by the replication and transcription complex (RTC). Here, we present a structural snapshot of SARS-CoV-2 RTC as it transitions toward cap structure synthesis. We determine the atomic cryo-EM structure of an extended RTC assembled by nsp7-nsp82-nsp12-nsp132-RNA and a single RNA-binding protein, nsp9. Nsp9 binds tightly to nsp12 (RdRp) NiRAN, allowing nsp9 N terminus inserting into the catalytic center of nsp12 NiRAN, which then inhibits activity. We also show that nsp12 NiRAN possesses guanylyltransferase activity, catalyzing the formation of cap core structure (GpppA). The orientation of nsp13 that anchors the 5' extension of template RNA shows a remarkable conformational shift, resulting in zinc finger 3 of its ZBD inserting into a minor groove of paired template-primer RNA. These results reason an intermediate state of RTC toward mRNA synthesis, pave a way to understand the RTC architecture, and provide a target for antiviral development.