Summary information and primary citation
- PDB-id
- 7d2l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (2.75 Å)
- Summary
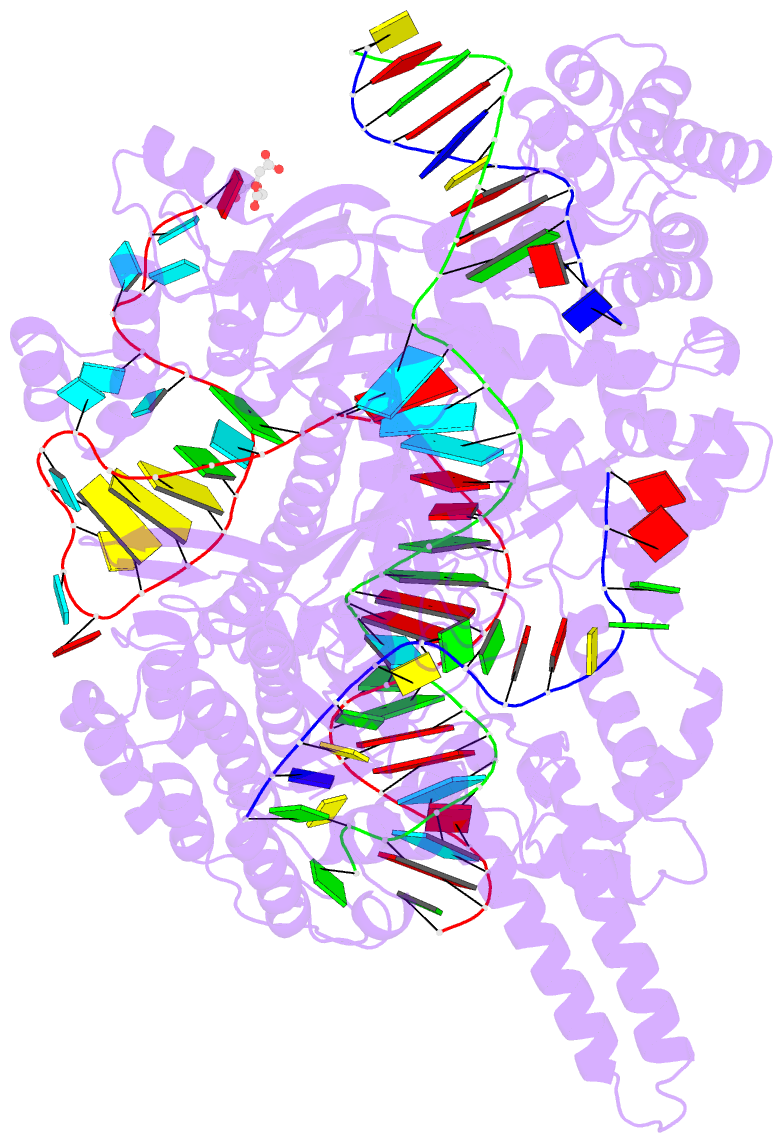
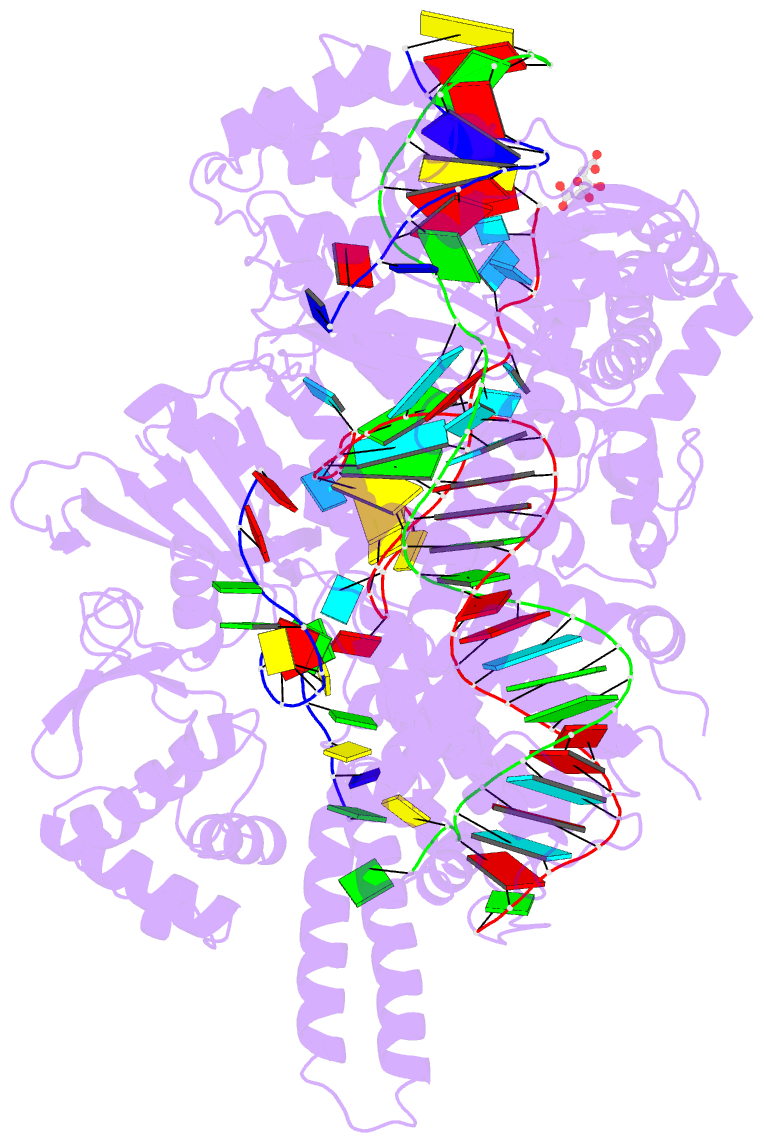
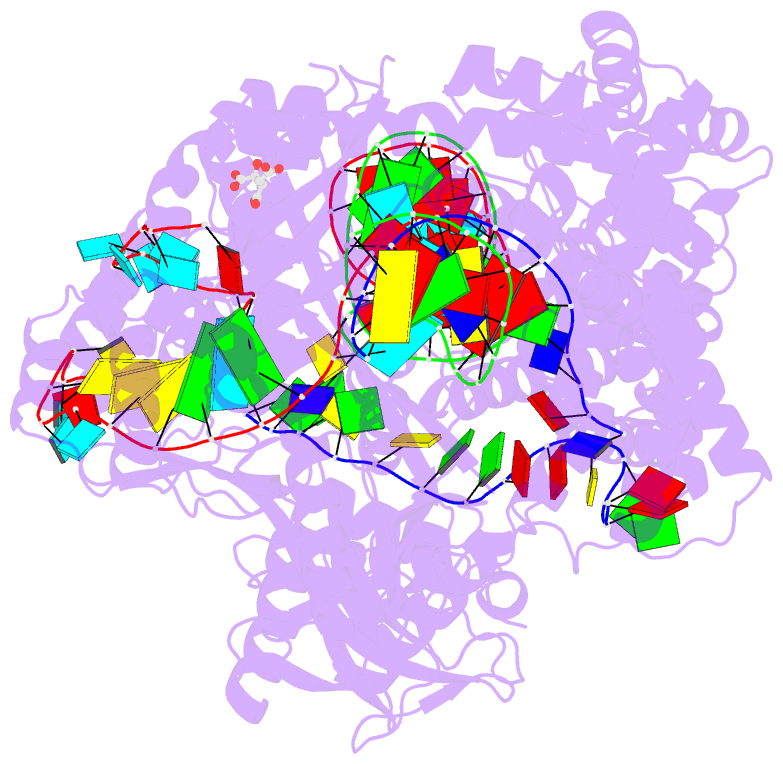
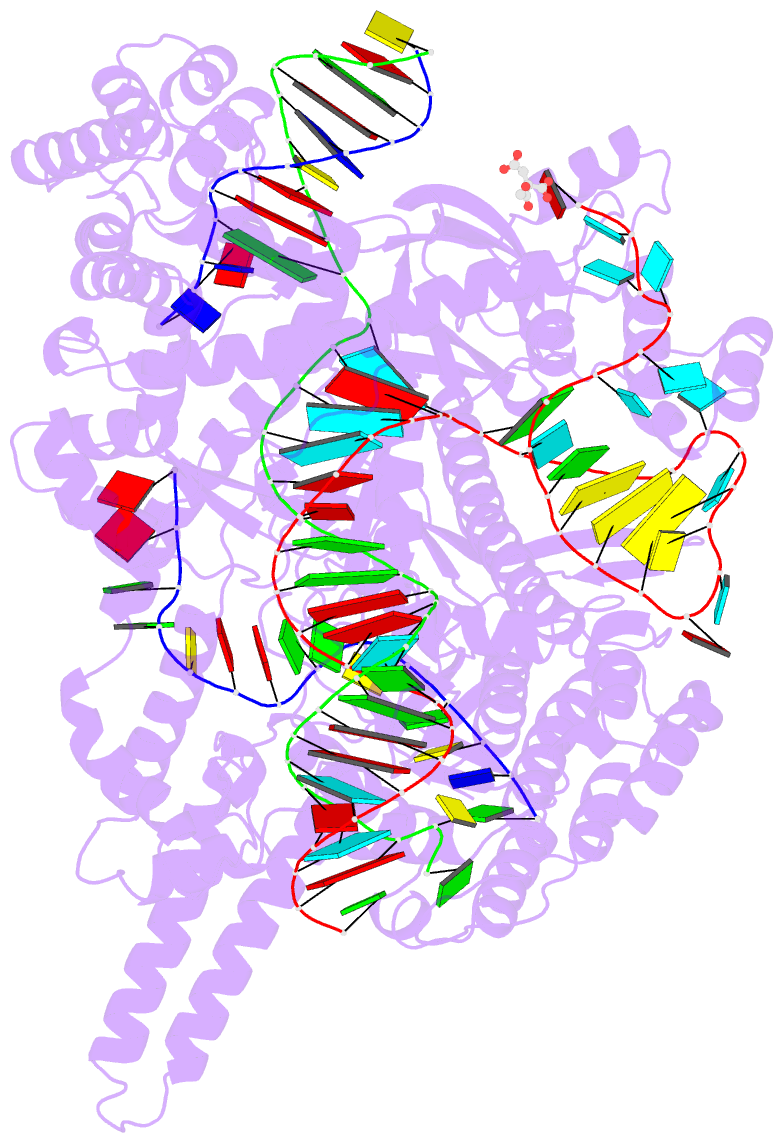
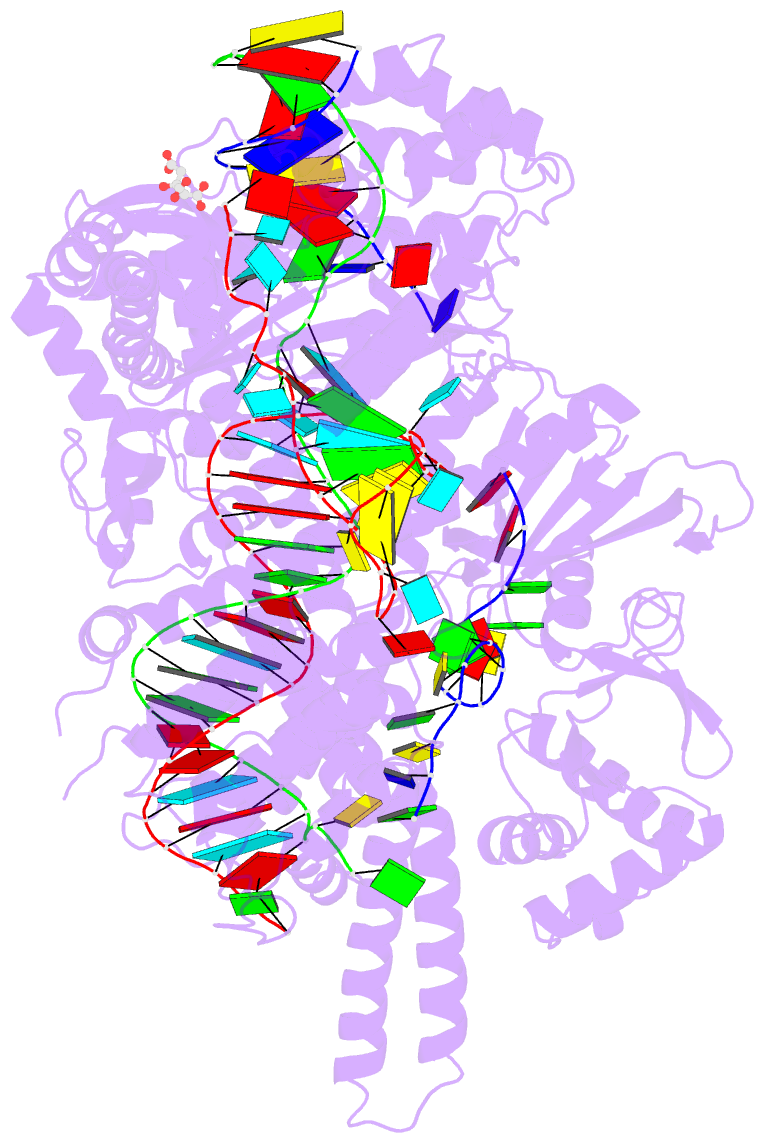
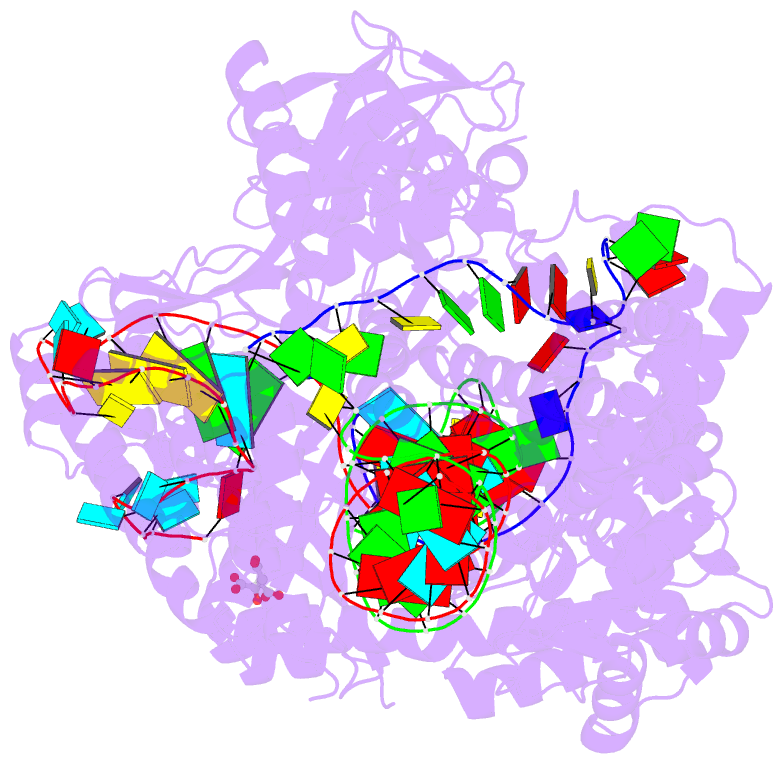
- Crystal structure of the cas12i1 r-loop complex before target DNA cleavage
- Reference
- Zhang B, Luo D, Li Y, Perculija V, Chen J, Lin J, Ye Y, Ouyang S (2021): "Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1." Nat Commun, 12, 3476. doi: 10.1038/s41467-021-23876-5.
- Abstract
- Cas12i is a newly identified member of the functionally diverse type V CRISPR-Cas effectors. Although Cas12i has the potential to serve as genome-editing tool, its structural and functional characteristics need to be investigated in more detail before effective application. Here we report the crystal structures of the Cas12i1 R-loop complexes before and after target DNA cleavage to elucidate the mechanisms underlying target DNA duplex unwinding, R-loop formation and cis cleavage. The structure of the R-loop complex after target DNA cleavage also provides information regarding trans cleavage. Besides, we report a crystal structure of the Cas12i1 binary complex interacting with a pseudo target oligonucleotide, which mimics target interrogation. Upon target DNA duplex binding, the Cas12i1 PAM-interacting cleft undergoes a remarkable open-to-closed adjustment. Notably, a zipper motif in the Helical-I domain facilitates unzipping of the target DNA duplex. Formation of the 19-bp crRNA-target DNA strand heteroduplex in the R-loop complexes triggers a conformational rearrangement and unleashes the DNase activity. This study provides valuable insights for developing Cas12i1 into a reliable genome-editing tool.