Summary information and primary citation
- PDB-id
- 7d3x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- NMR
- Summary
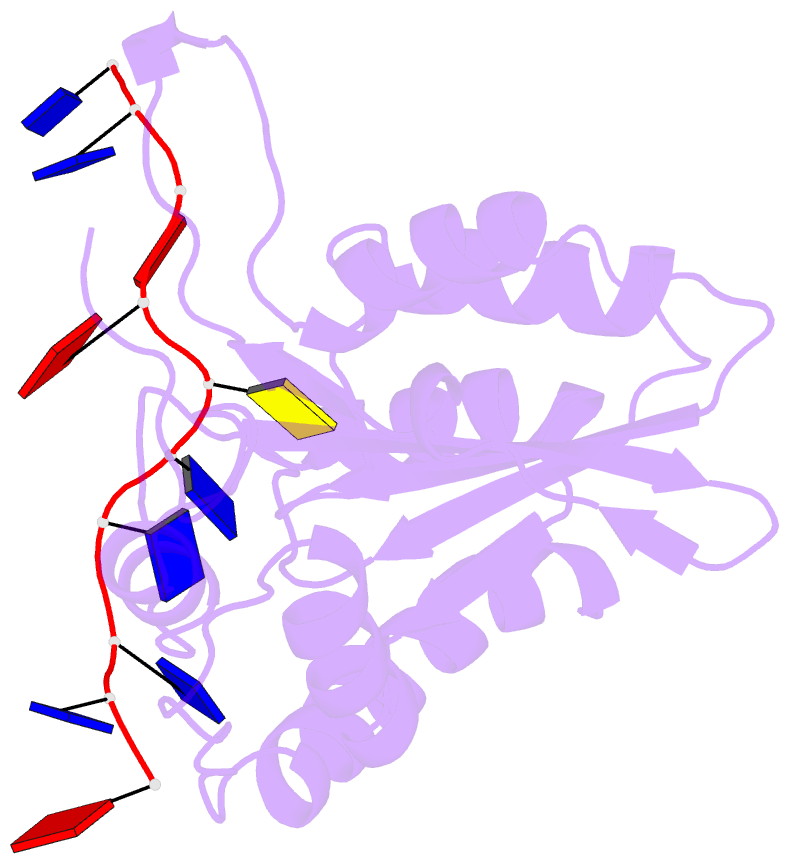
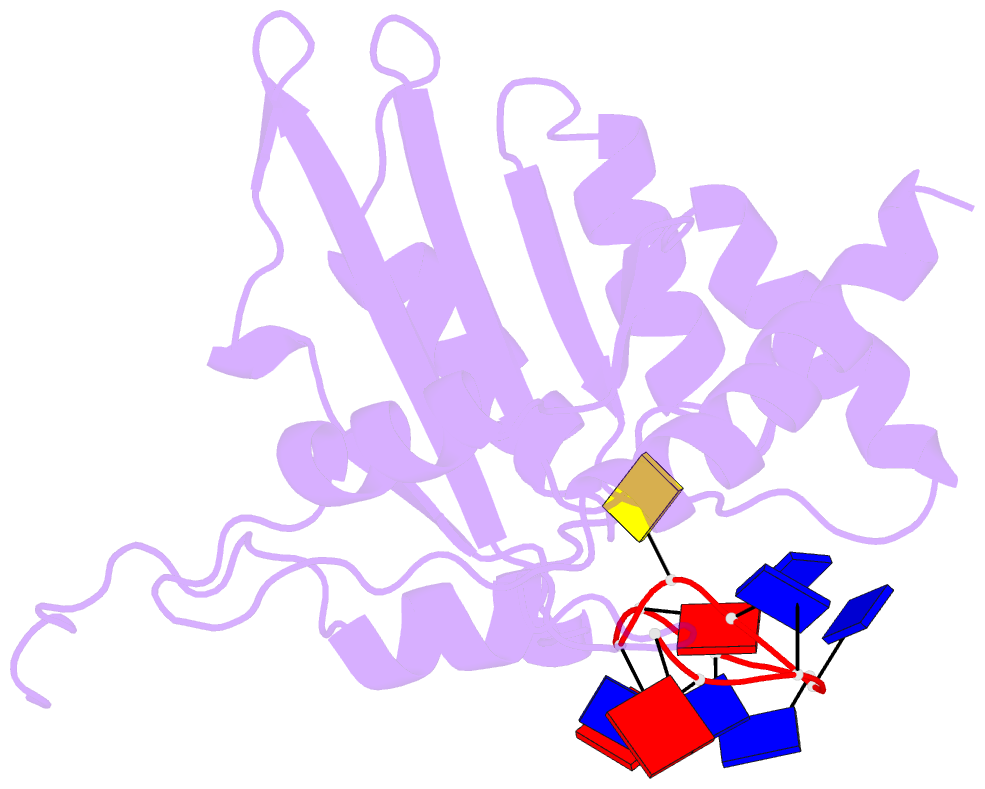
- Non-specific and specific interactions work cooperatively to promote cytidine deamination catalyzed by apobec3a
- Reference
- Liu Y, Lan W, Wang C, Cao C (2022): "Two different kinds of interaction modes of deaminase APOBEC3A with single-stranded DNA in solution detected by nuclear magnetic resonance." Protein Sci., 31, 443-453. doi: 10.1002/pro.4242.
- Abstract
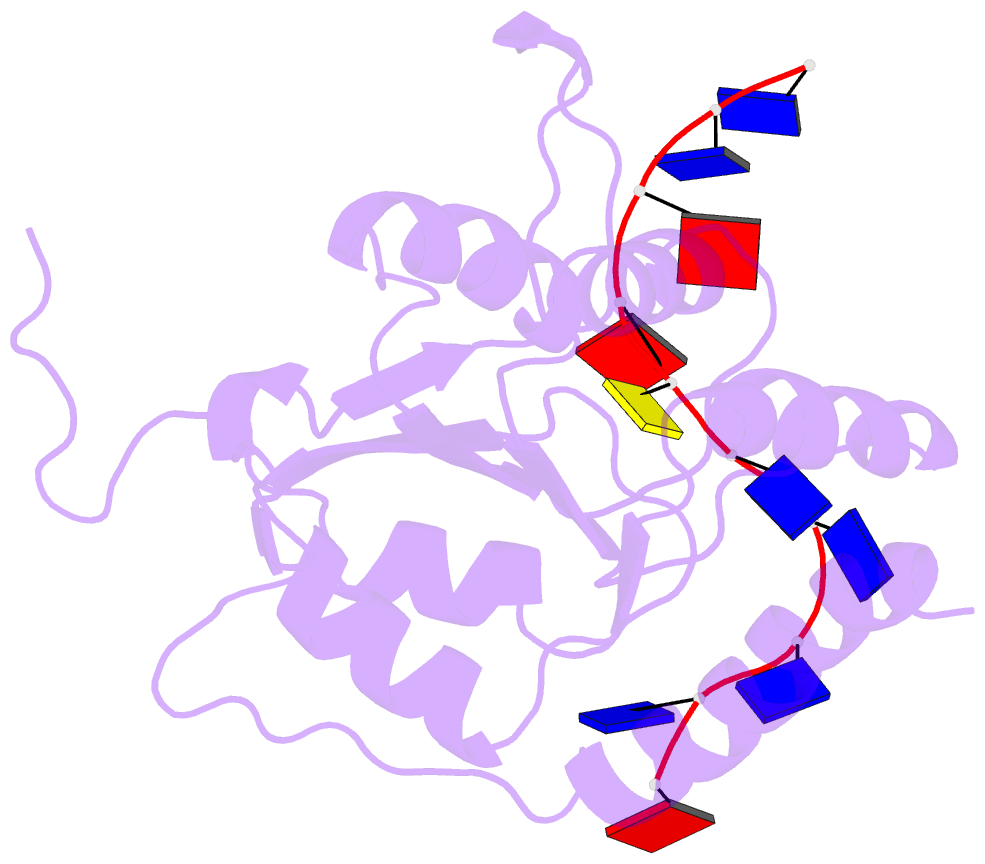
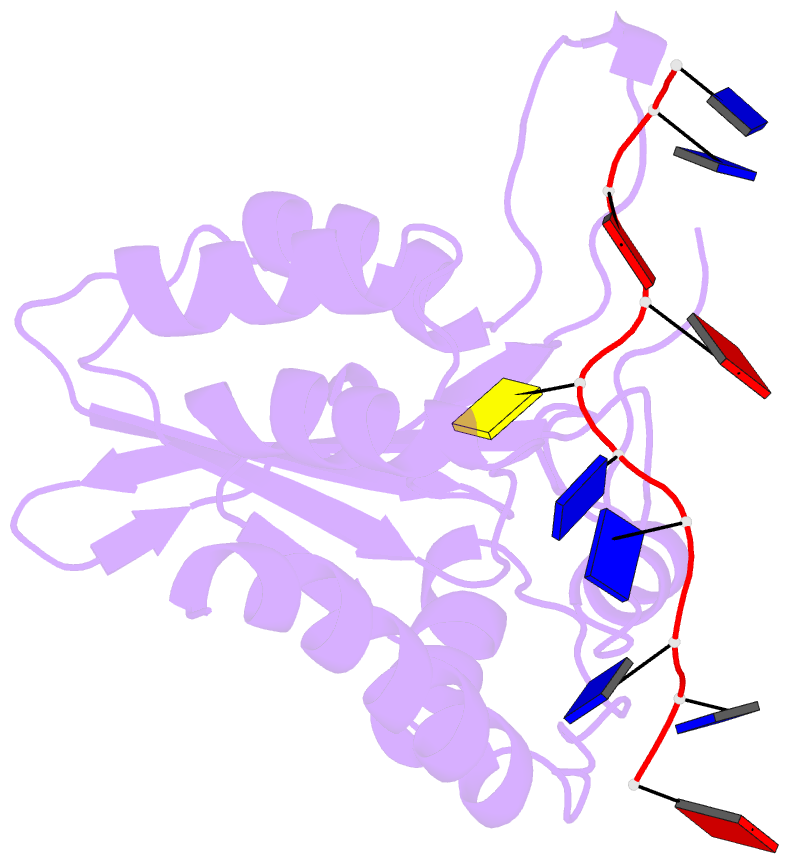
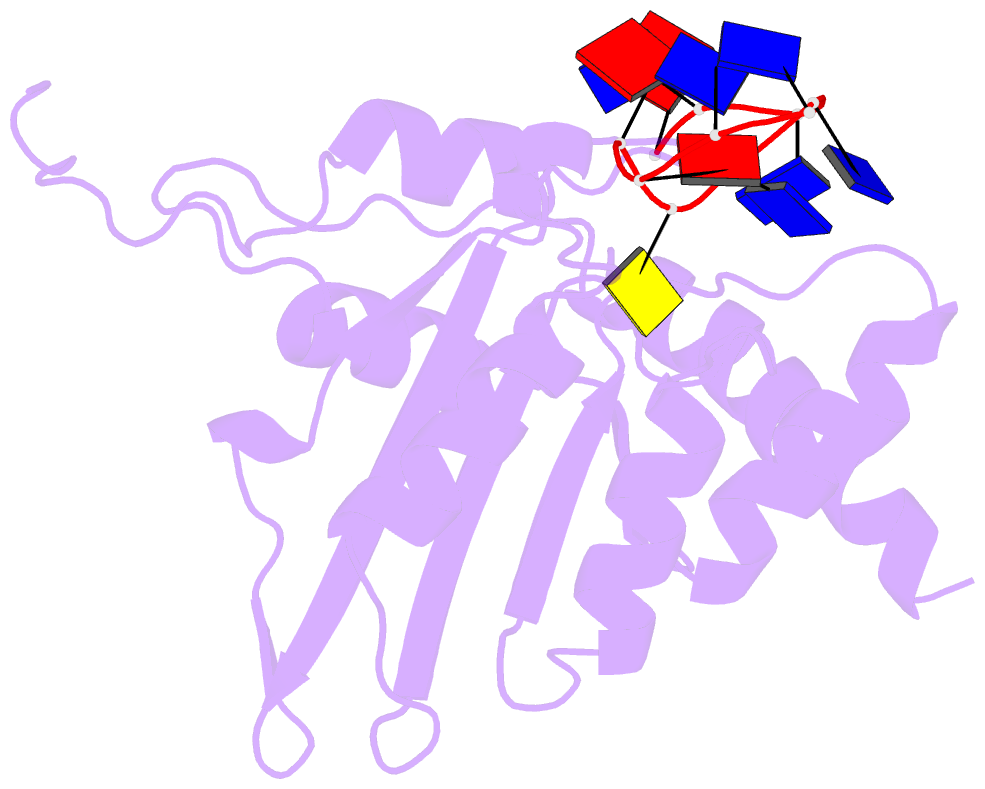
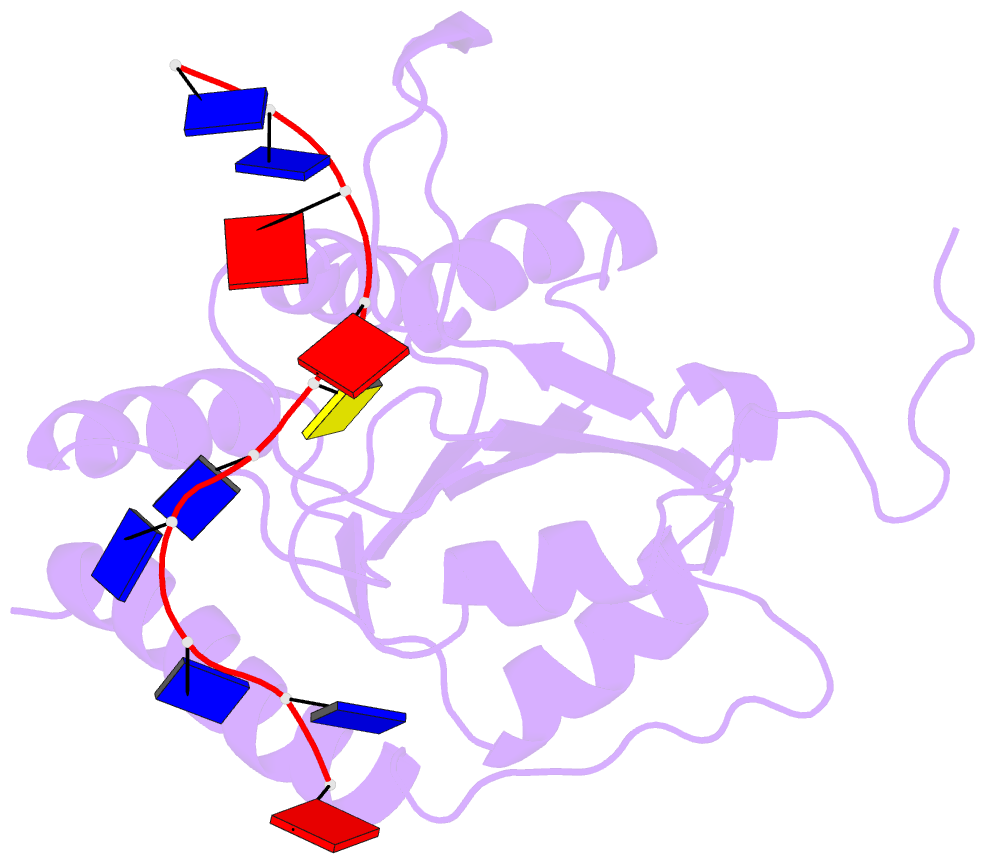
- APOBEC3A (A3A) deaminates deoxycytidine in target motif TC in a single-stranded DNA (we termed it as TC DNA), which mortally mutates viral pathogens and immunoglobulins, and leads to the diversification and lethality of cancers. The crystal structure of A3A-DNA revealed a unique U-shaped recognition mode of target base dC0 . However, when TC DNA was titrated into 15 N-labeled A3A solution, we observed two sets of 1 H-15 N cross-peaks of A3A in HSQC spectra, and two sets of 1 H-1 H cross-peaks of DNA in two-dimensional 13 C,15 N-filtered TOCSY spectra, indicating two different kinds of conformers of either A3A or TC DNA existing in solution. Here, mainly by NMR, we demonstrated that one DNA conformer interacted with one A3A conformer, forming a specific complex A3AS -DNAS in a way almost similar to that observed in the reported crystal A3A-DNA structure, where dC0 inserted into zinc ion binding center. While the other DNA conformer bound with another A3A conformer, but dC0 did not extend into the zinc-binding pocket, forming a nonspecific A3ANS -DNANS complex. The NMR solution structure implied three sites Asn61 , His182 and Arg189 were necessary to DNA recognition. These observations indicate a distinctive way from that reported in X-ray crystal structure, suggesting an unexpected mode of deaminase APOBEC3A to identify target motif TC in DNA in solution.