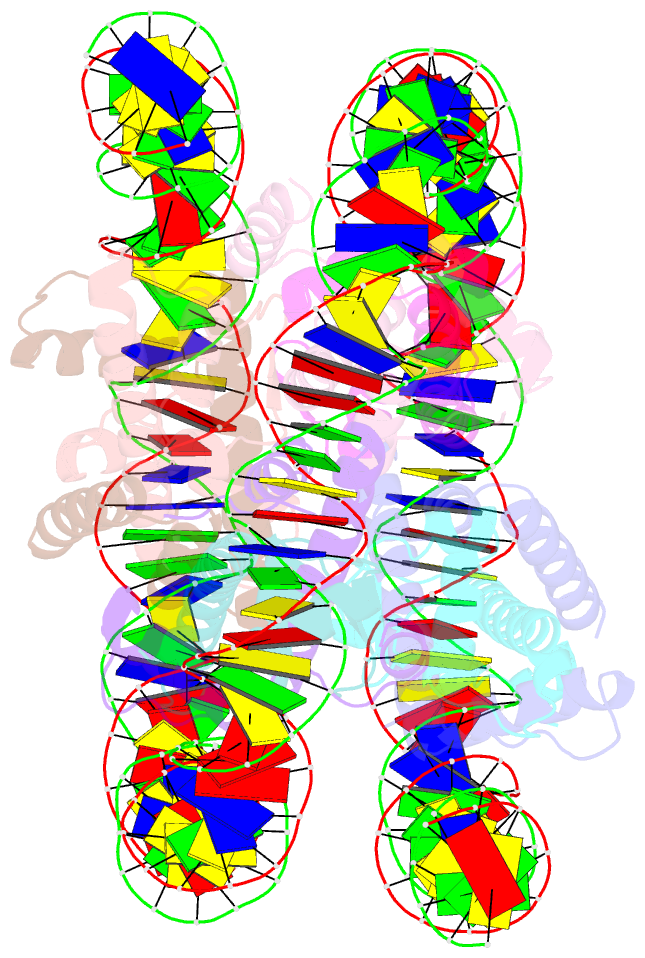
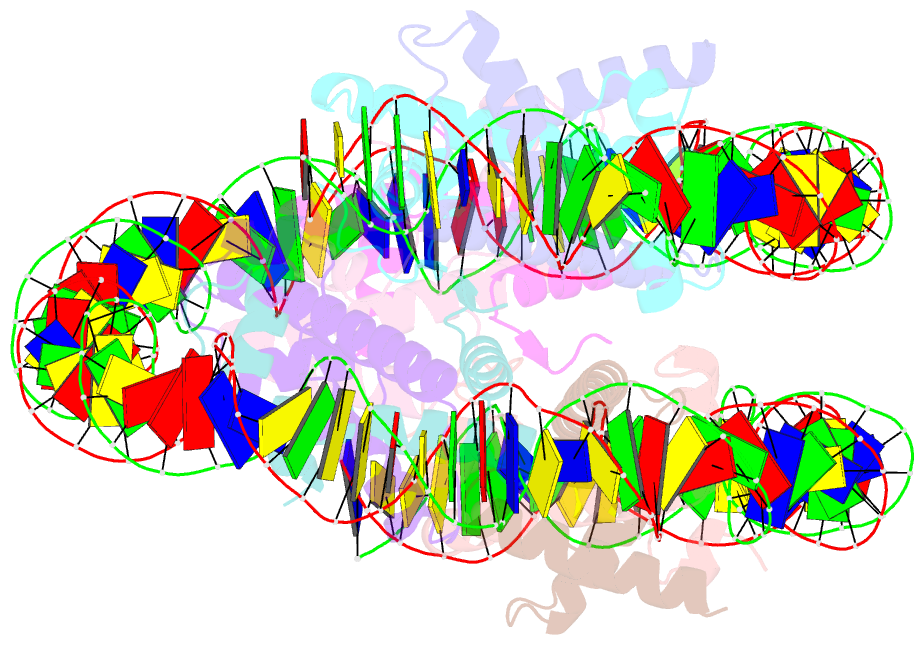
Summary information and primary citation
- PDB-id
- 7d69; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (3.57 Å)
- Summary
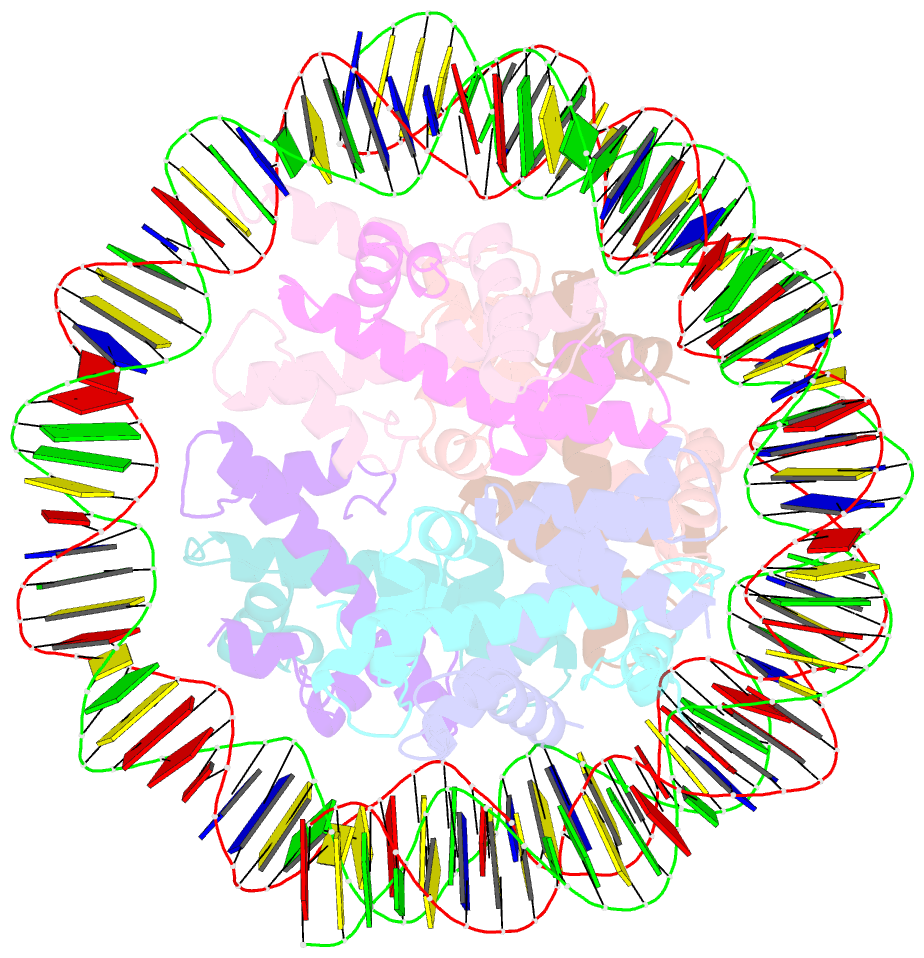
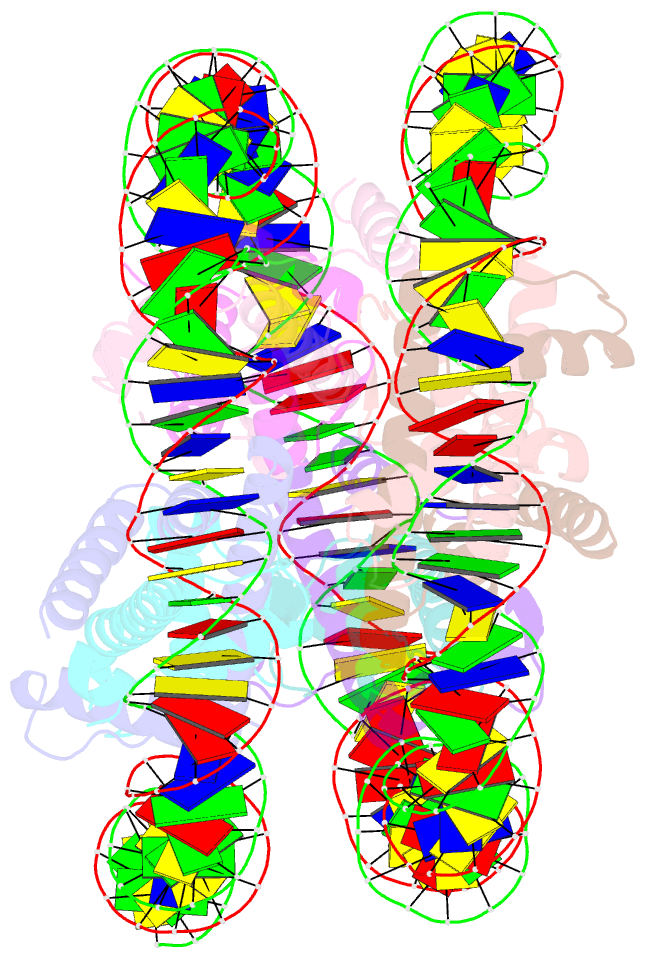
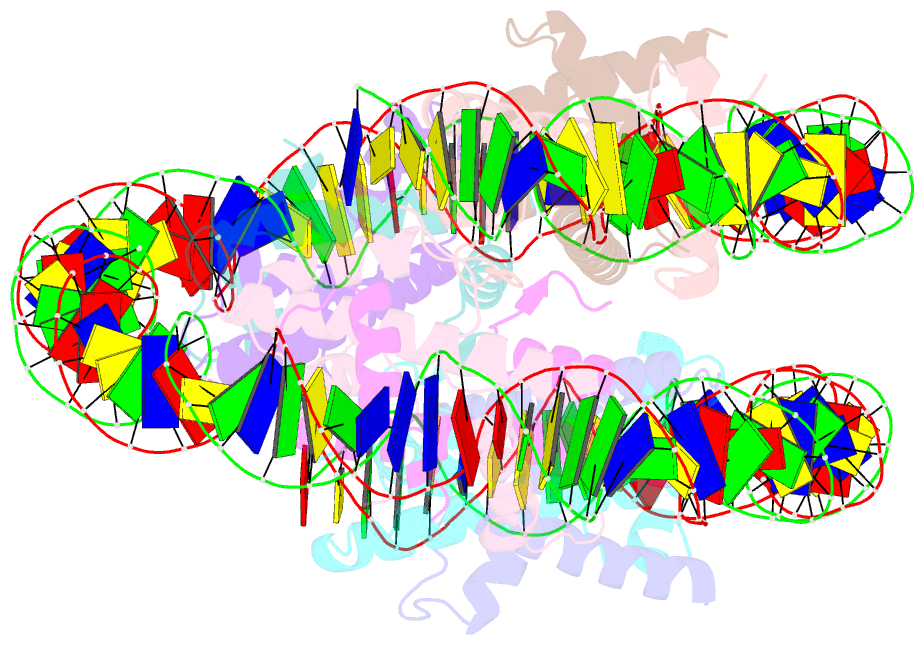
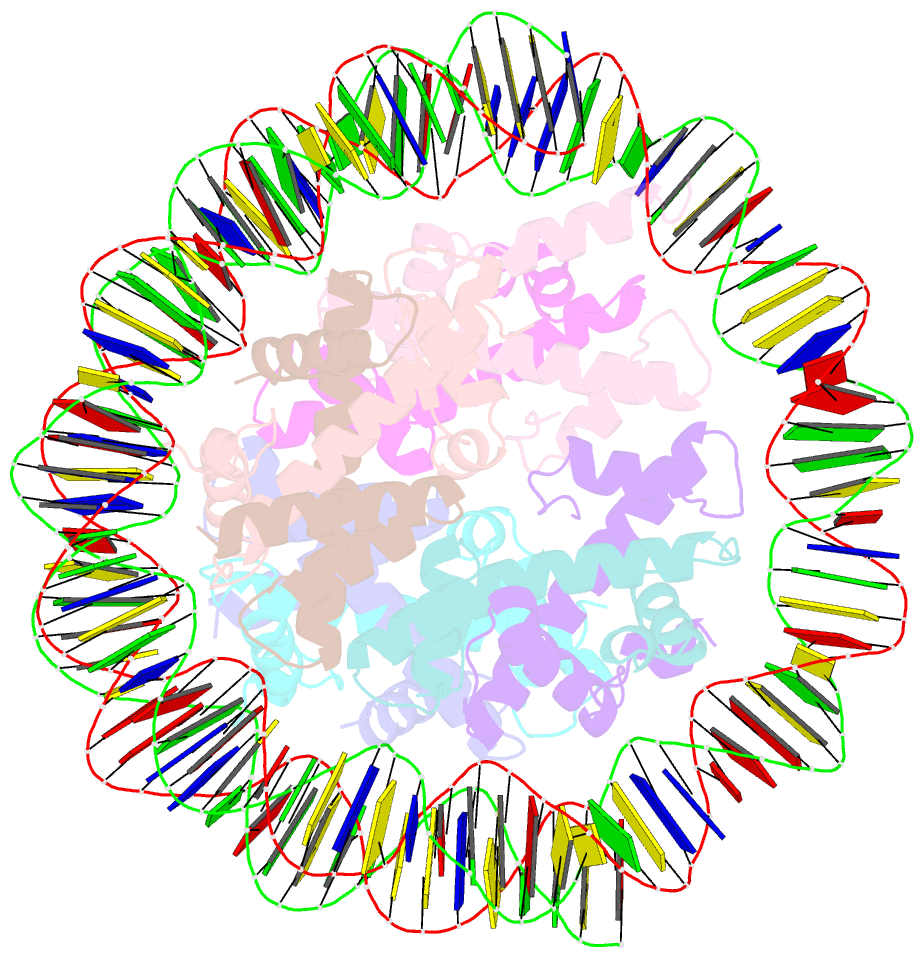
- cryo-EM structure of the nucleosome containing giardia histones
- Reference
- Sato S, Takizawa Y, Hoshikawa F, Dacher M, Tanaka H, Tachiwana H, Kujirai T, Iikura Y, Ho CH, Adachi N, Patwal I, Flaus A, Kurumizaka H (2021): "Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones." Nucleic Acids Res., 49, 8934-8946. doi: 10.1093/nar/gkab644.
- Abstract
- Giardia lamblia is a pathogenic unicellular eukaryotic parasite that causes giardiasis. Its genome encodes the canonical histones H2A, H2B, H3, and H4, which share low amino acid sequence identity with their human orthologues. We determined the structure of the G. lamblia nucleosome core particle (NCP) at 3.6 Å resolution by cryo-electron microscopy. G. lamblia histones form a characteristic NCP, in which the visible 125 base-pair region of the DNA is wrapped in a left-handed supercoil. The acidic patch on the G. lamblia octamer is deeper, due to an insertion extending the H2B α1 helix and L1 loop, and thus cannot bind the LANA acidic patch binding peptide. The DNA and histone regions near the DNA entry-exit sites could not be assigned, suggesting that these regions are asymmetrically flexible in the G. lamblia NCP. Characterization by thermal unfolding in solution revealed that both the H2A-H2B and DNA association with the G. lamblia H3-H4 were weaker than those for human H3-H4. These results demonstrate the uniformity of the histone octamer as the organizing platform for eukaryotic chromatin, but also illustrate the unrecognized capability for large scale sequence variations that enable the adaptability of histone octamer surfaces and confer internal stability.