Summary information and primary citation
- PDB-id
- 7d7c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.6 Å)
- Summary
- Cryoem structure of gp55-dependent RNA polymerase-promoter open complex
- Reference
- Shi J, Wen A, Jin S, Gao B, Huang Y, Feng Y (2021): "Transcription activation by a sliding clamp." Nat Commun, 12, 1131. doi: 10.1038/s41467-021-21392-0.
- Abstract
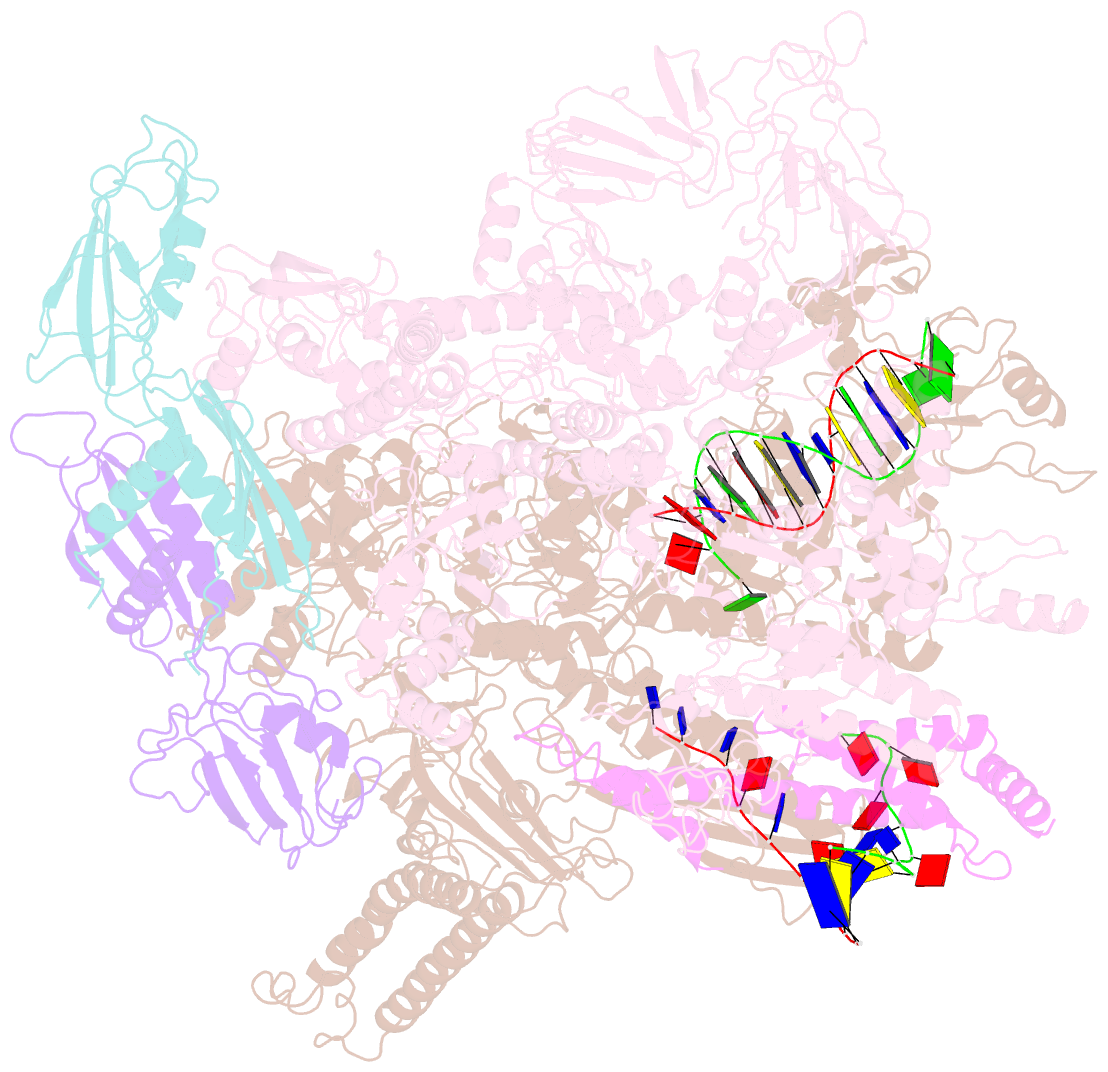
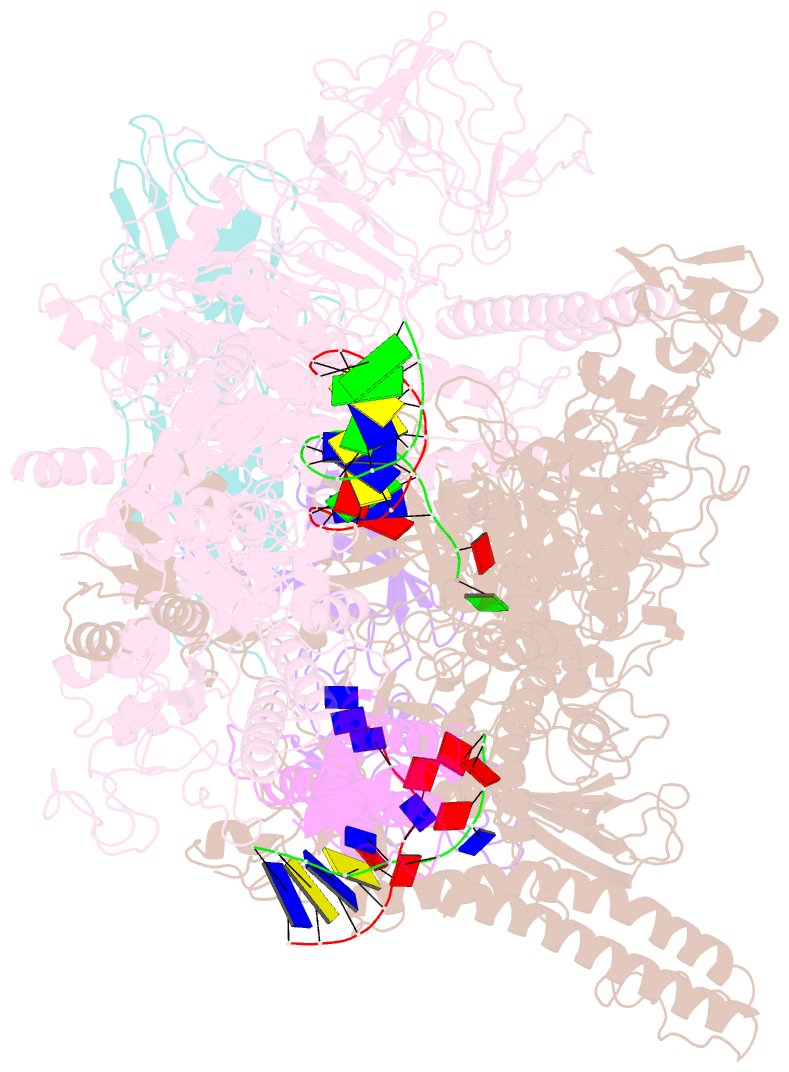
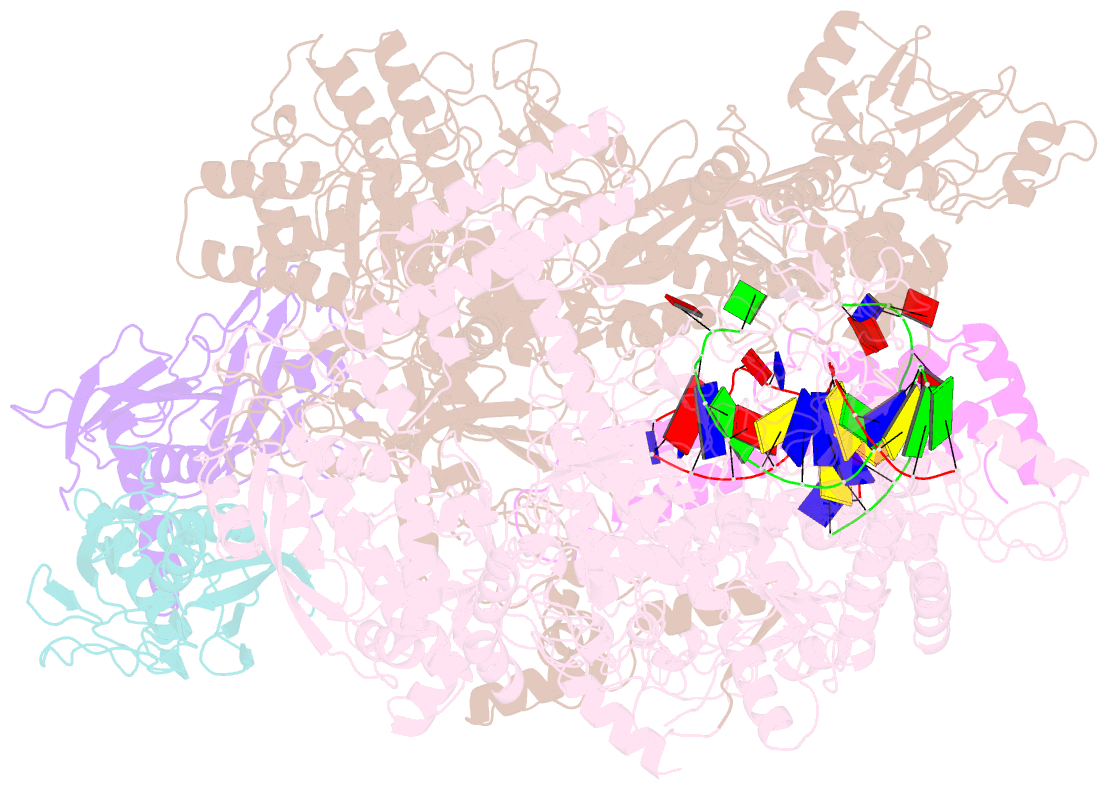
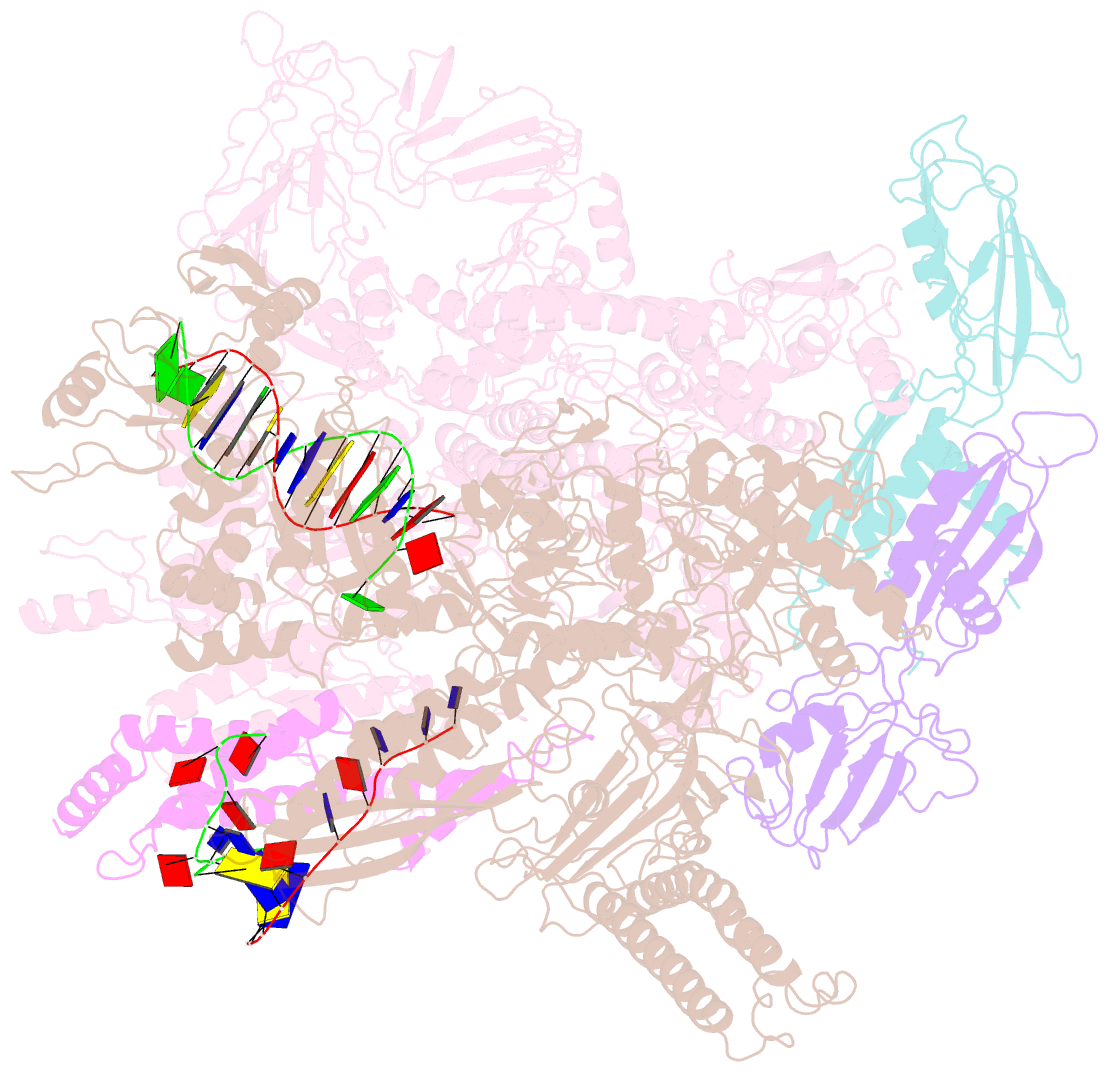
- Transcription activation of bacteriophage T4 late genes is accomplished by a transcription activation complex containing RNA polymerase (RNAP), the promoter specificity factor gp55, the coactivator gp33, and a universal component of cellular DNA replication, the sliding clamp gp45. Although genetic and biochemical studies have elucidated many aspects of T4 late gene transcription, no precise structure of the transcription machinery in the process is available. Here, we report the cryo-EM structures of a gp55-dependent RNAP-promoter open complex and an intact gp45-dependent transcription activation complex. The structures reveal the interactions between gp55 and the promoter DNA that mediate the recognition of T4 late promoters. In addition to the σR2 homology domain, gp55 has a helix-loop-helix motif that chaperons the template-strand single-stranded DNA of the transcription bubble. Gp33 contacts both RNAP and the upstream double-stranded DNA. Gp45 encircles the DNA and tethers RNAP to it, supporting the idea that gp45 switches the promoter search from three-dimensional diffusion mode to one-dimensional scanning mode.