Summary information and primary citation
- PDB-id
- 7dkg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (3.0 Å)
- Summary
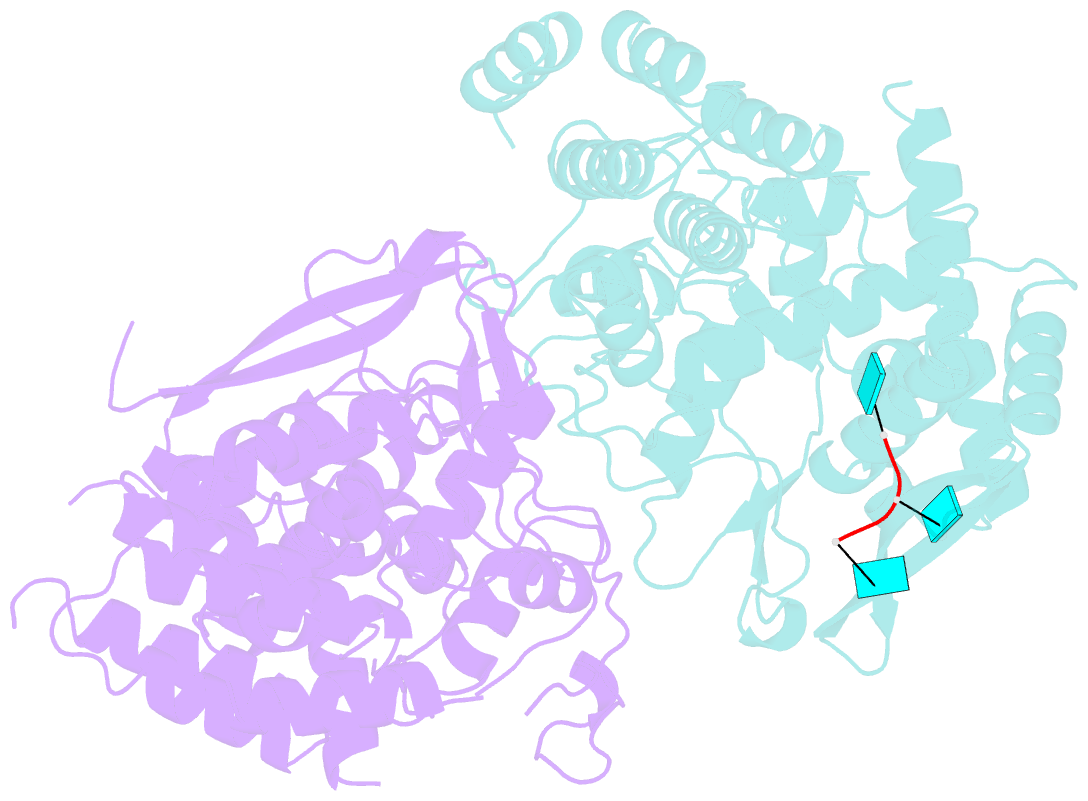
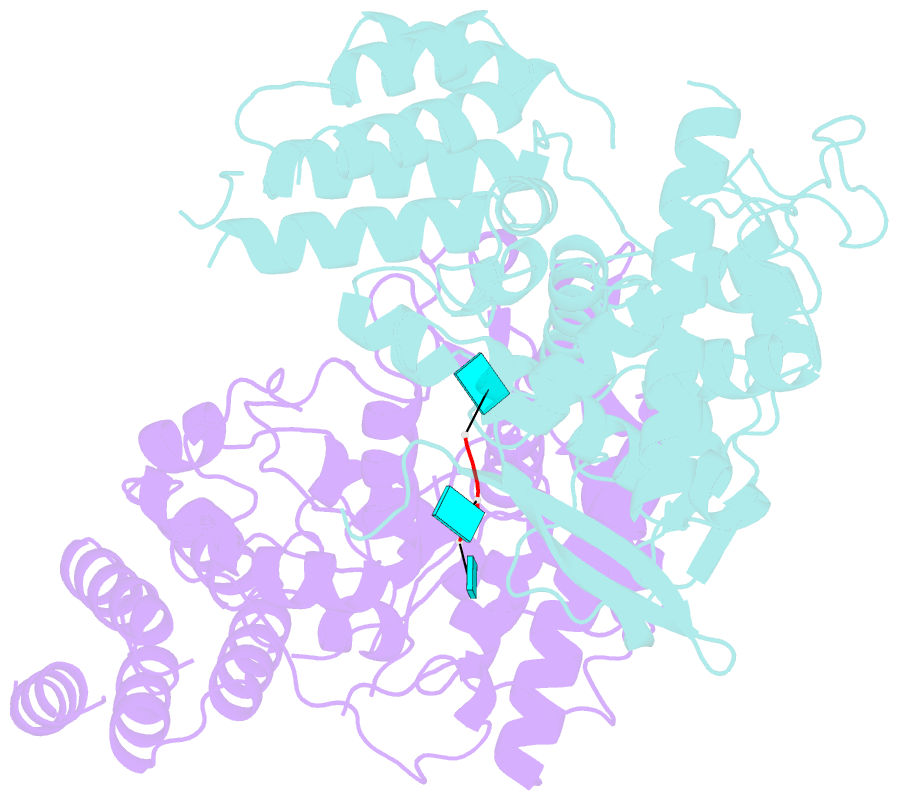
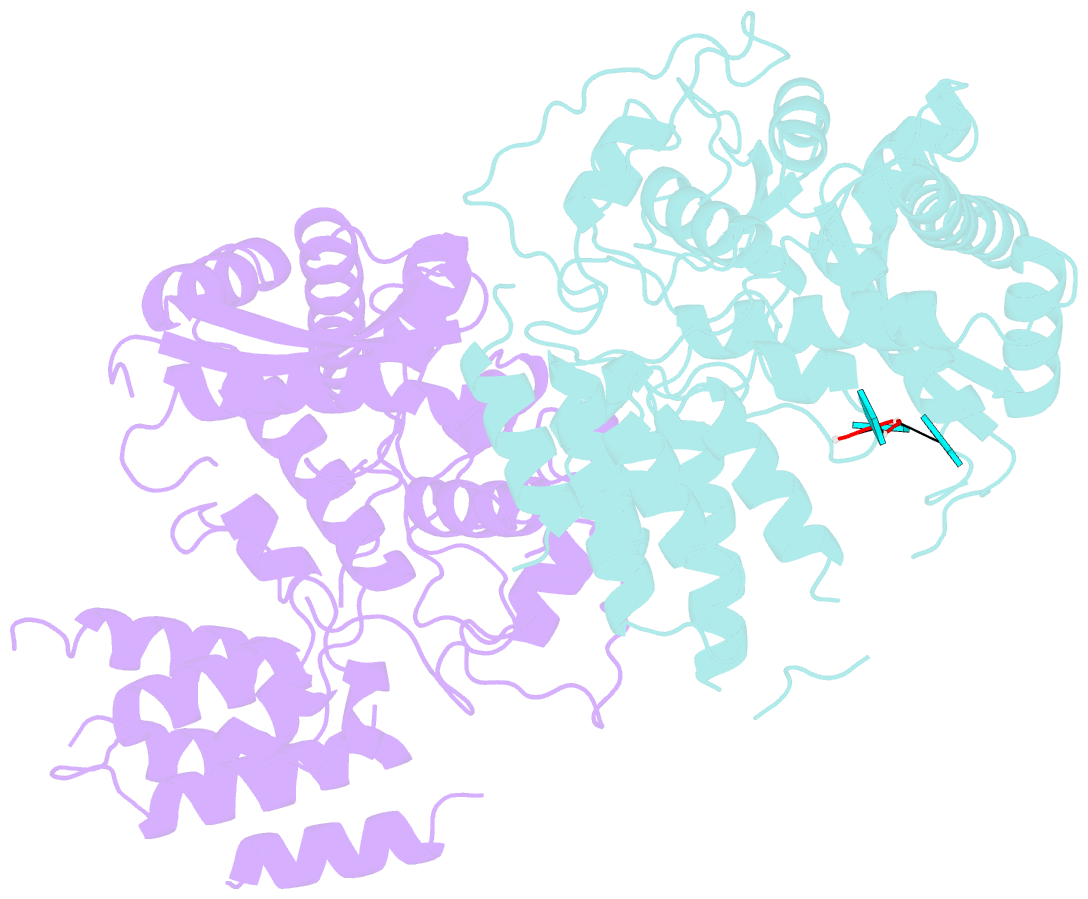
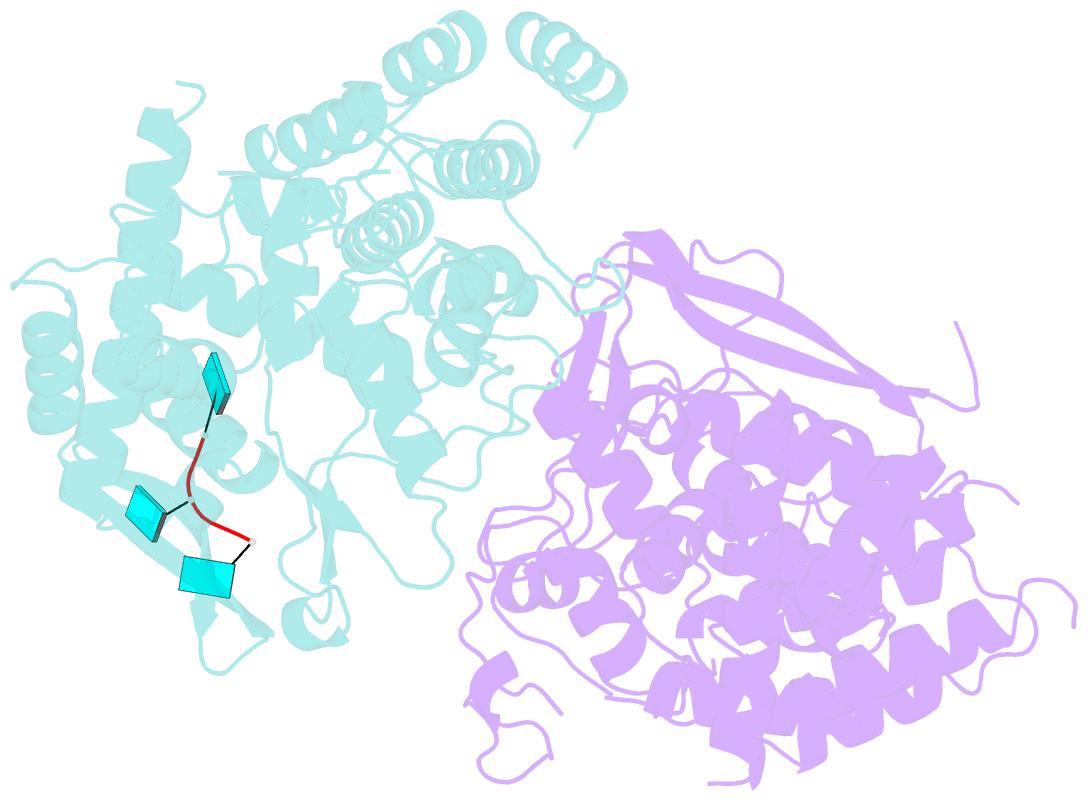
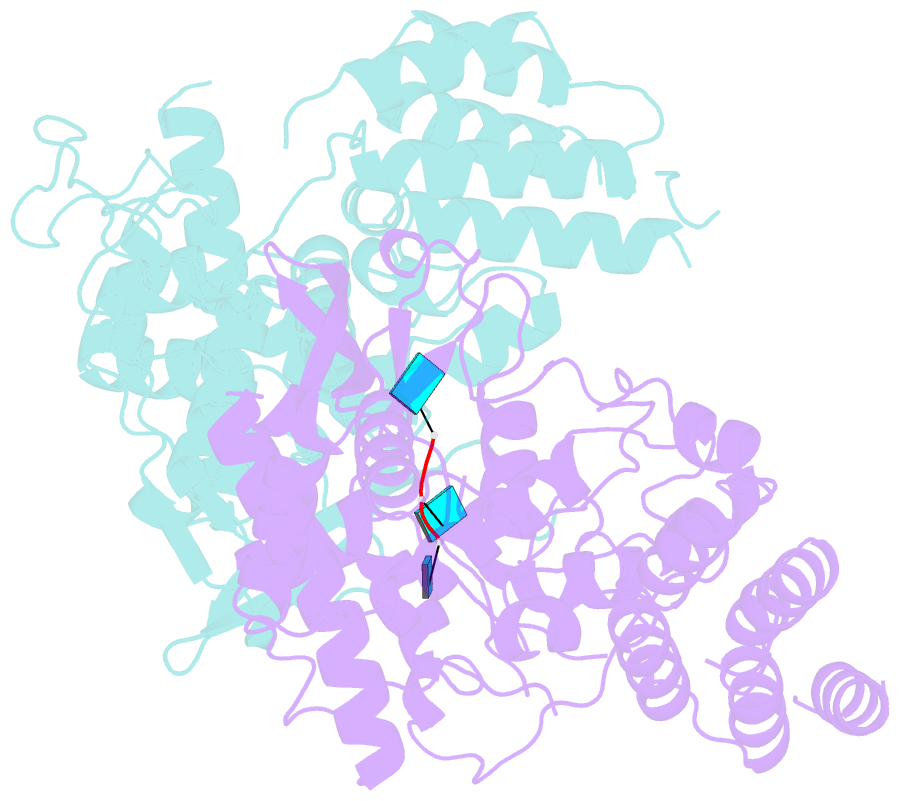
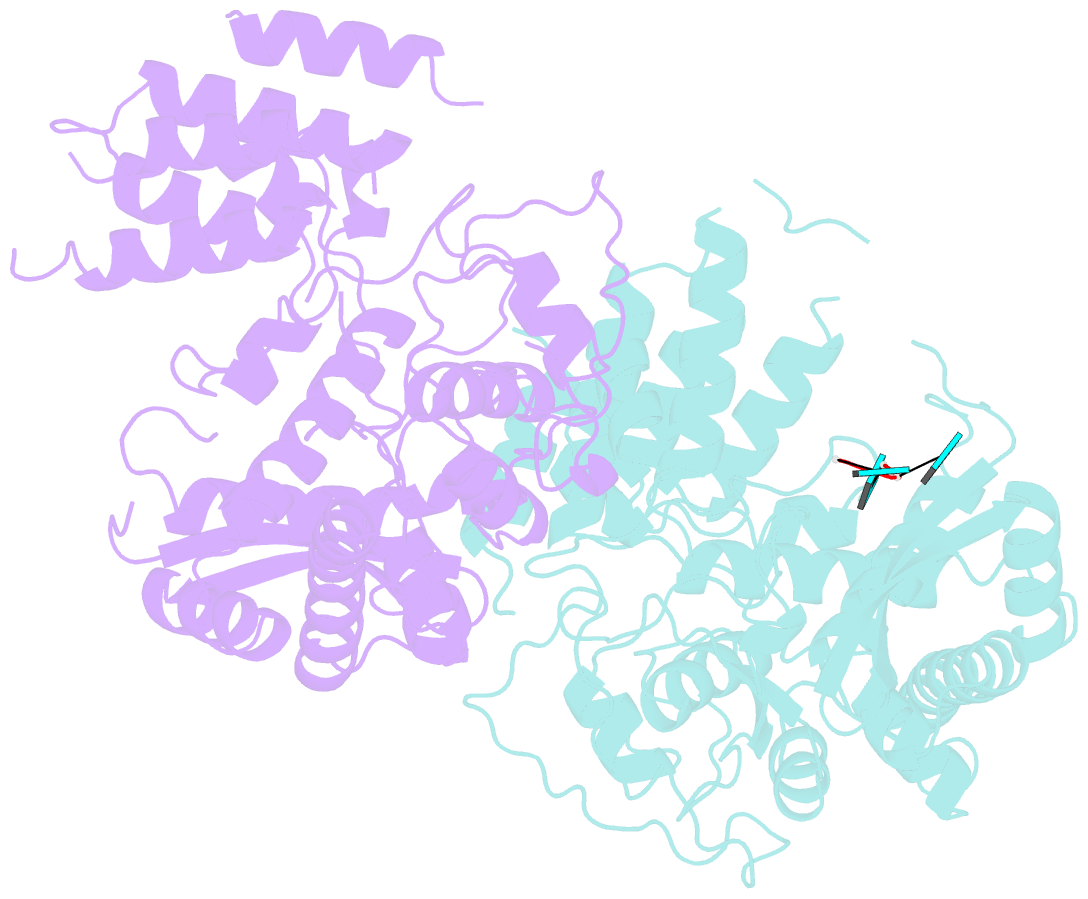
- Influenza h5n1 nucleoprotein (truncated) in complex with nucleotides
- Reference
- Tang YS, Xu S, Chen YW, Wang JH, Shaw PC (2021): "Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction." Nucleic Acids Res., 49, 4144-4154. doi: 10.1093/nar/gkab203.
- Abstract
- The nucleoprotein (NP) of influenza virus is the core component of the ribonucleoprotein (RNP) and performs multiple structural and functional roles. Structures of the influenza A, B and D NP molecules have been solved previously, but structural information on how NP interacts with RNA remains elusive. Here we present the crystal structure of an obligate monomer of H5N1 NP in complex with RNA nucleotides to 2.3 Å, and a C-terminal truncation of this mutant, also in complex with RNA nucleotides, to 3 Å. In both structures, three nucleotides were identified near two positive grooves of NP suggested to be important for RNA binding. Structural evidence supports that conformational changes of flexible loops and the C-terminal tail both play important roles in the binding of RNA. Based on the structure, we propose a mechanism by which NP captures RNA by flexible loops and transfers it onto the positive binding grooves. Binding of RNA by NP is a crucial step for template re-encapsidation during transcription and replication and cRNP formation. Our structures thus provide insights into the molecular virology of the influenza virus.