Summary information and primary citation
- PDB-id
- 7dmq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-RNA
- Method
- cryo-EM (3.06 Å)
- Summary
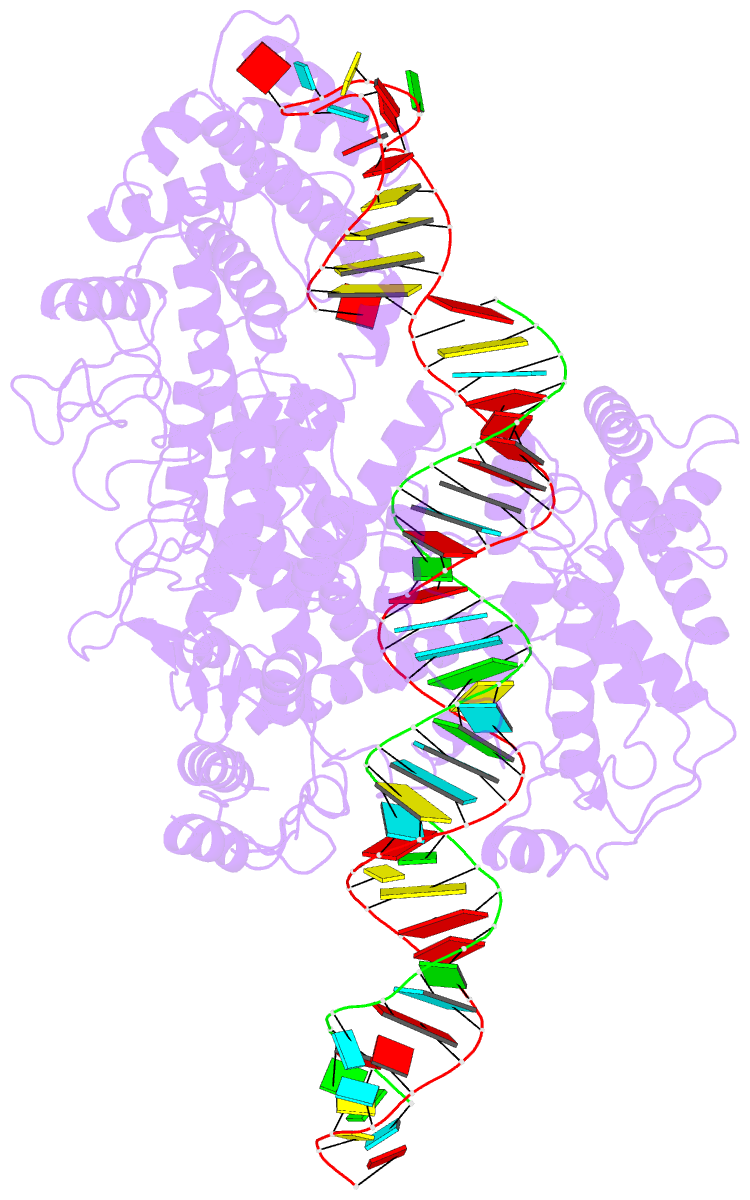
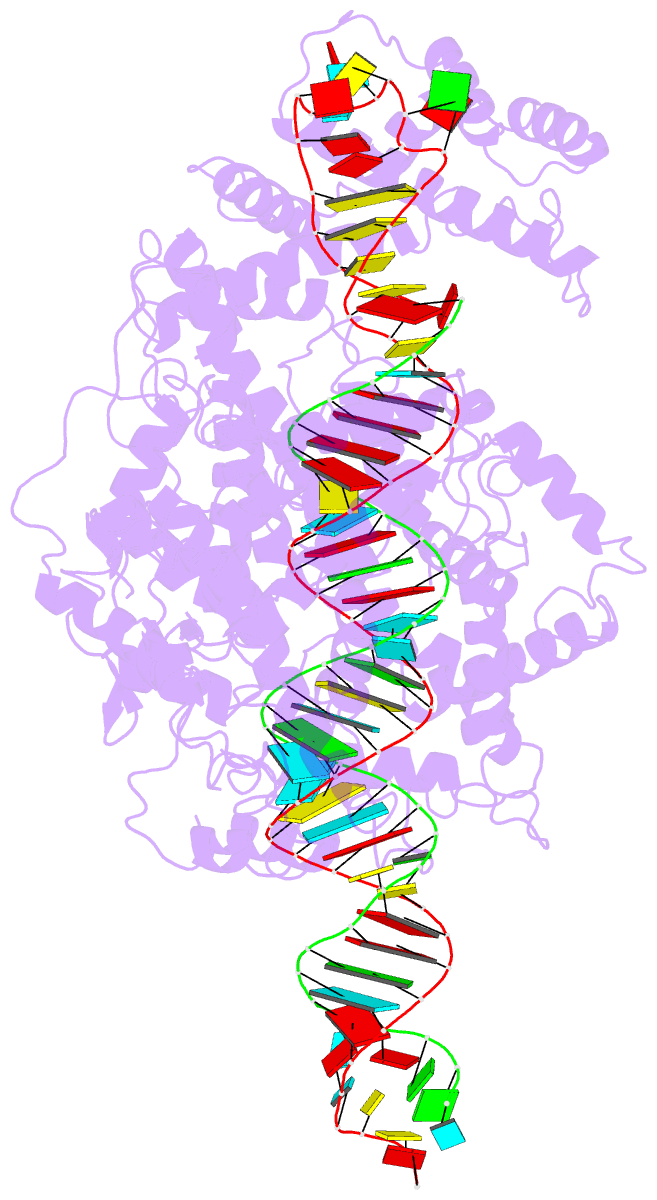
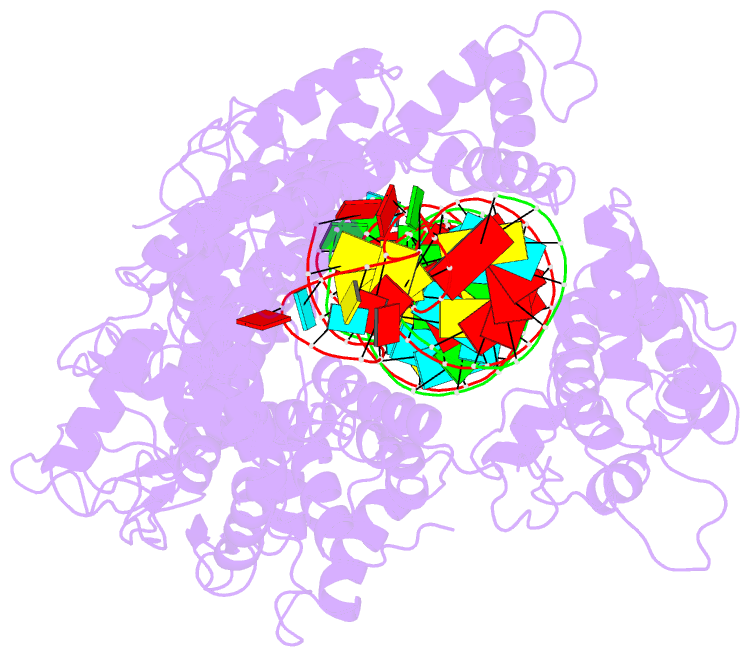
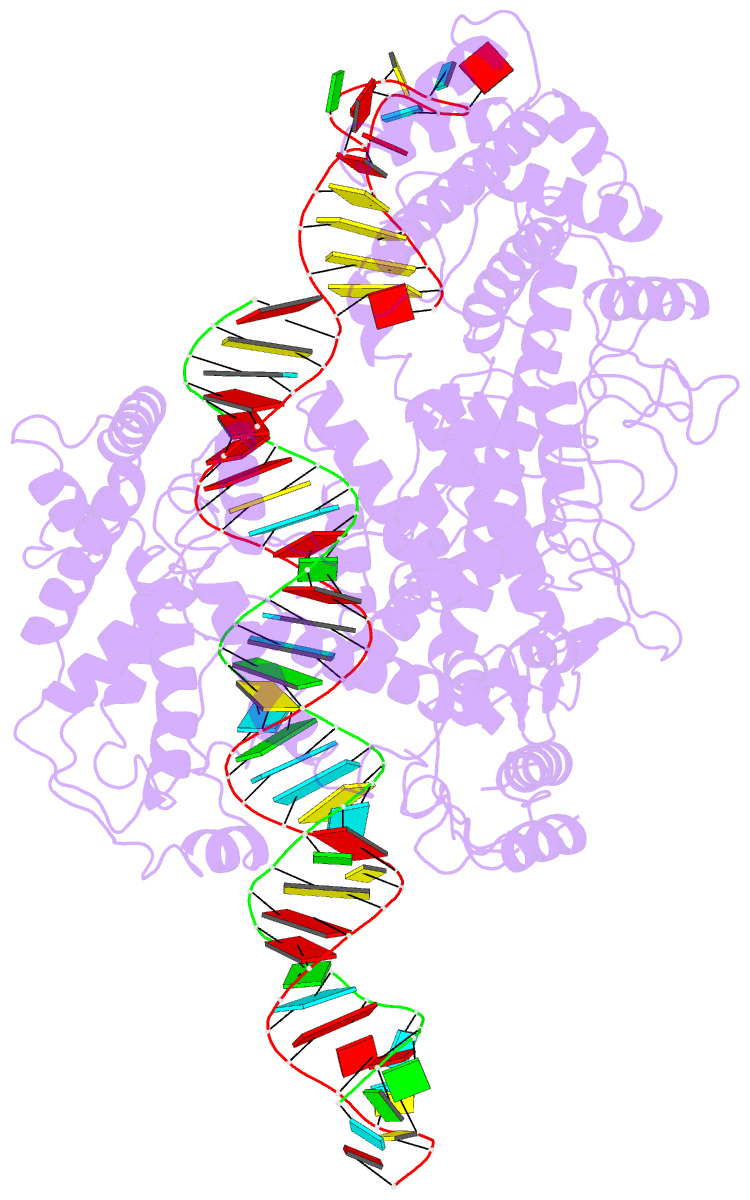
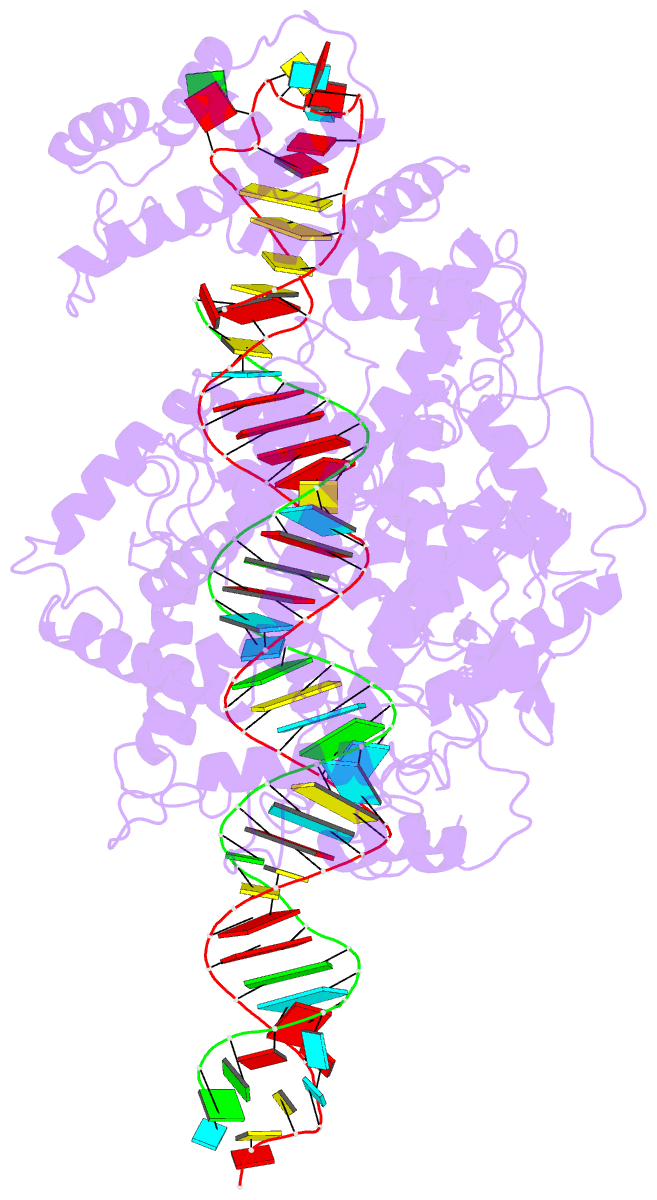
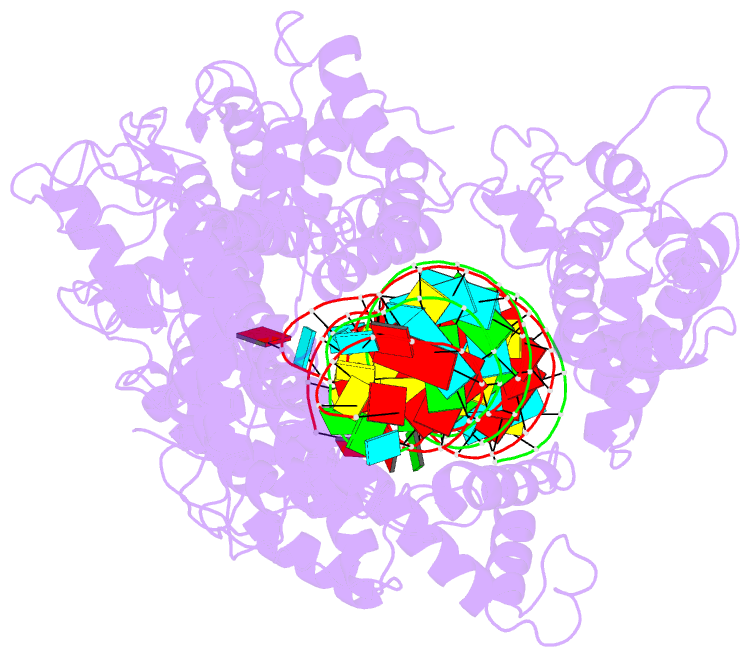
- cryo-EM structure of lshcas13a-crrna-anti-tag RNA complex
- Reference
- Wang B, Zhang T, Yin J, Yu Y, Xu W, Ding J, Patel DJ, Yang H (2021): "Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems." Mol.Cell, 81, 1100. doi: 10.1016/j.molcel.2020.12.033.
- Abstract
- Bacteria and archaea apply CRISPR-Cas surveillance complexes to defend against foreign invaders. These invading genetic elements are captured and integrated into the CRISPR array as spacer elements, guiding sequence-specific DNA/RNA targeting and cleavage. Recently, in vivo studies have shown that target RNAs with extended complementarity with repeat sequences flanking the target element (tag:anti-tag pairing) can dramatically reduce RNA cleavage by the type VI-A Cas13a system. Here, we report the cryo-EM structure of Leptotrichia shahii LshCas13acrRNA in complex with target RNA harboring tag:anti-tag pairing complementarity, with the observed conformational changes providing a molecular explanation for inactivation of the composite HEPN domain cleavage activity. These structural insights, together with in vitro biochemical and in vivo cell-based assays on key mutants, define the molecular principles underlying Cas13a's capacity to target and discriminate between self and non-self RNA targets. Our studies illuminate approaches to regulate Cas13a's cleavage activity, thereby influencing Cas13a-mediated biotechnological applications.