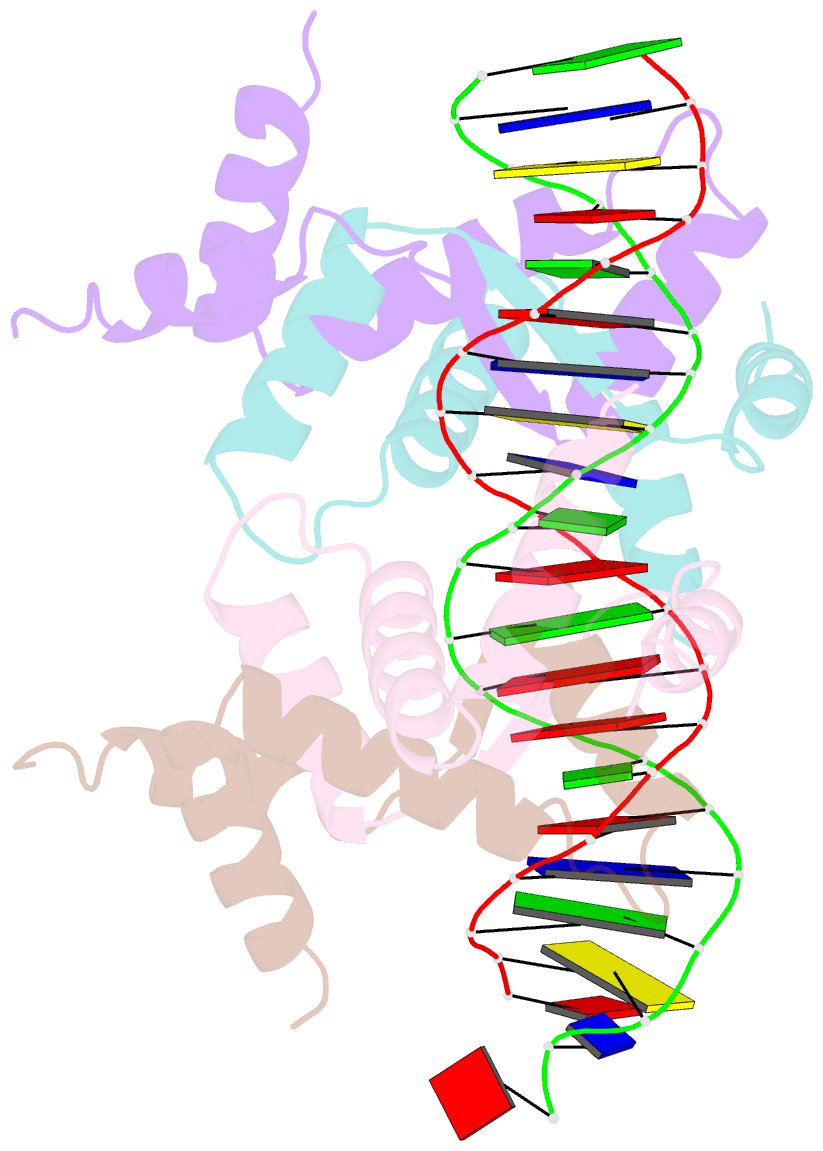
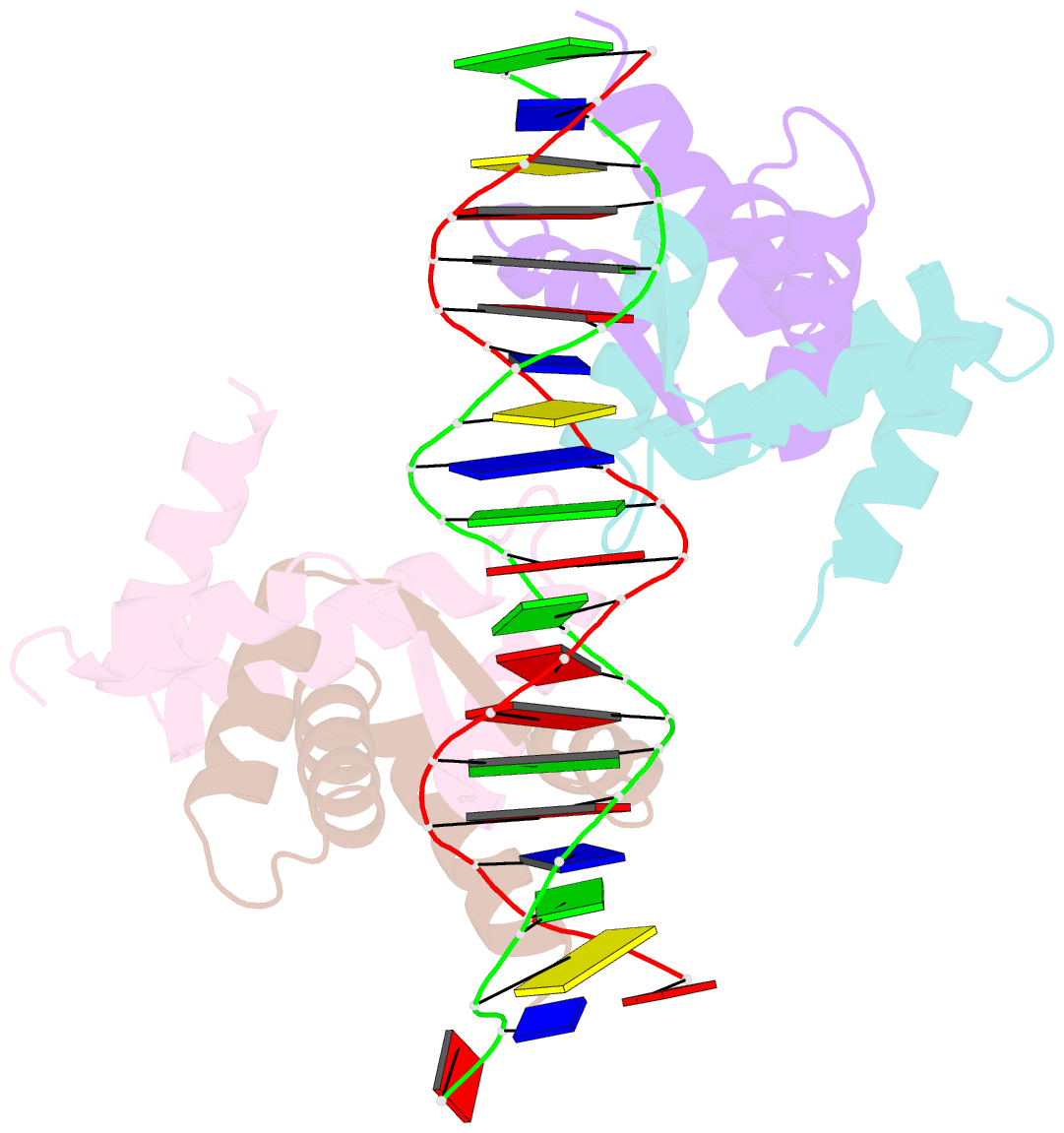
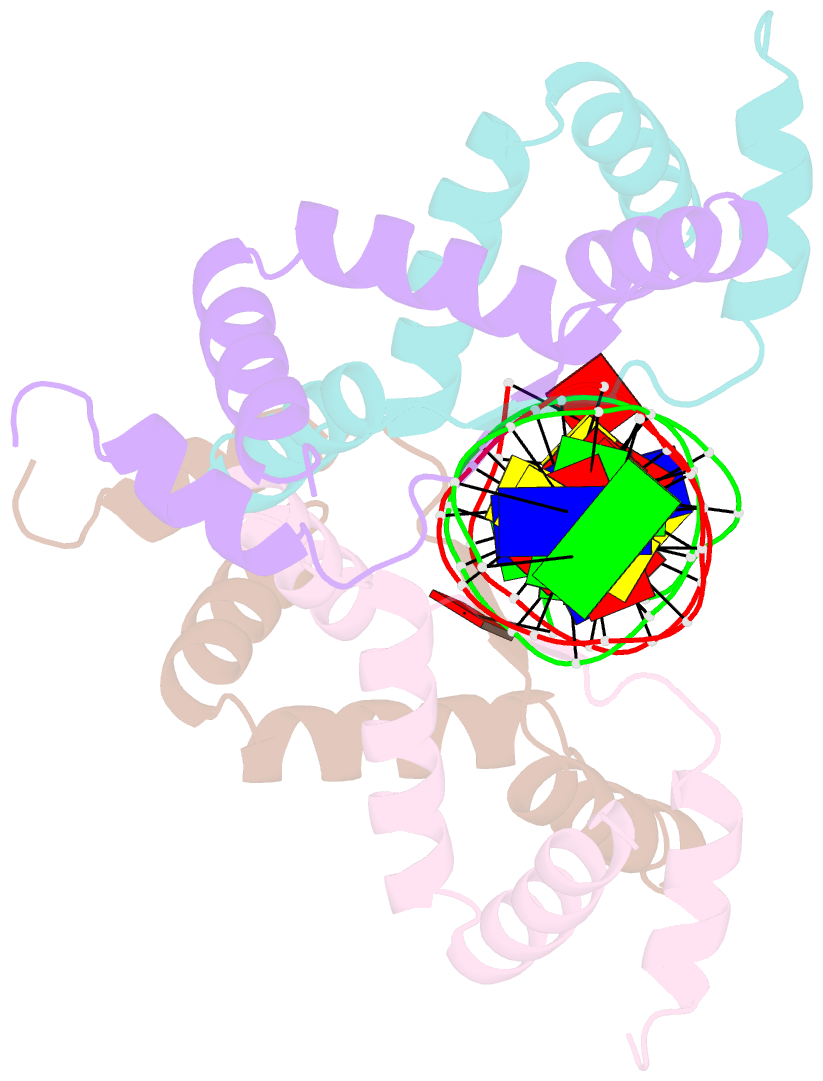
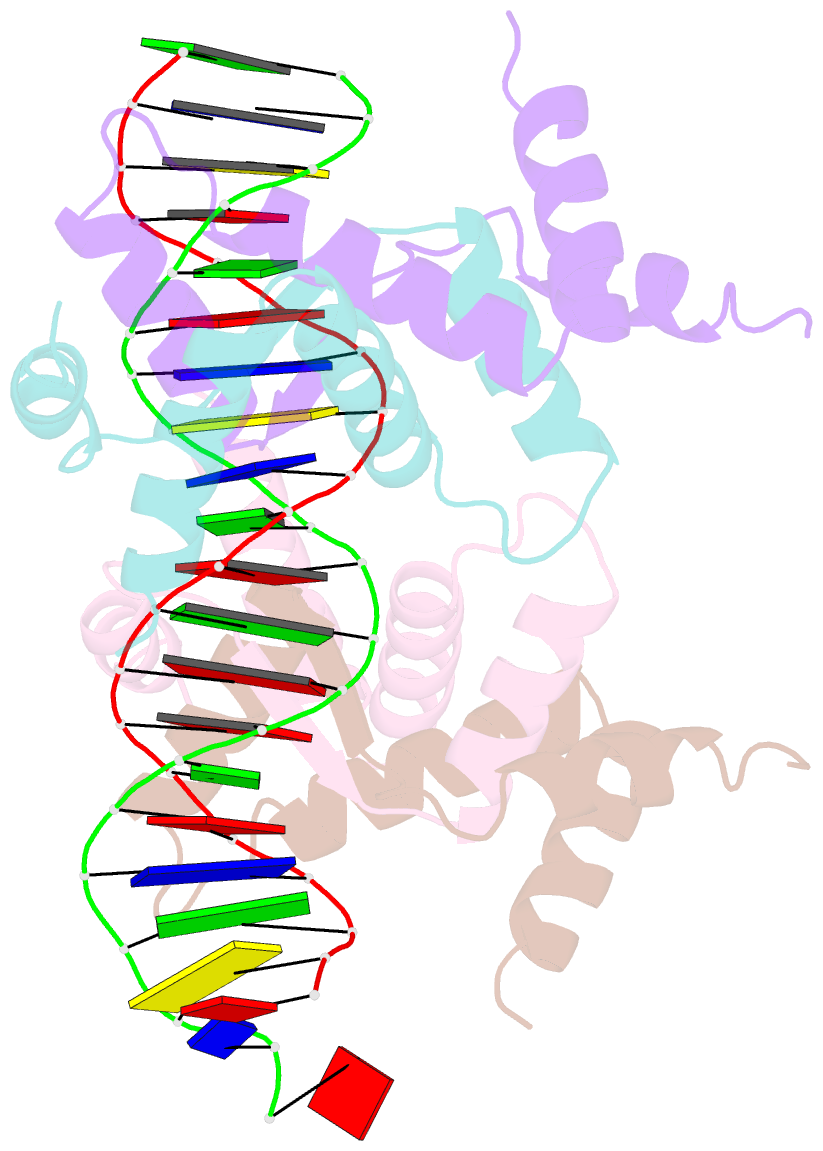
Summary information and primary citation
- PDB-id
- 7dv2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (3.1 Å)
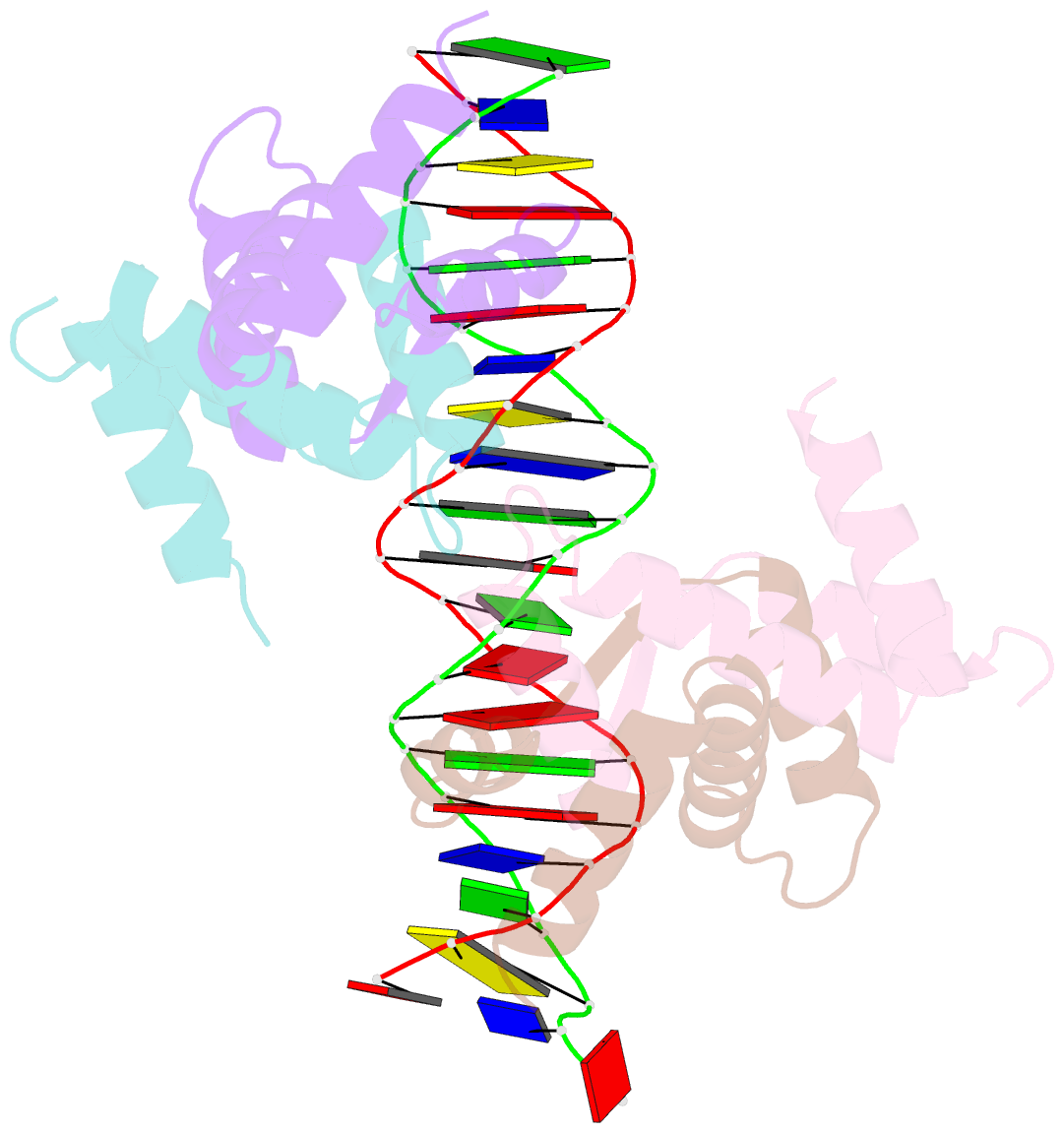
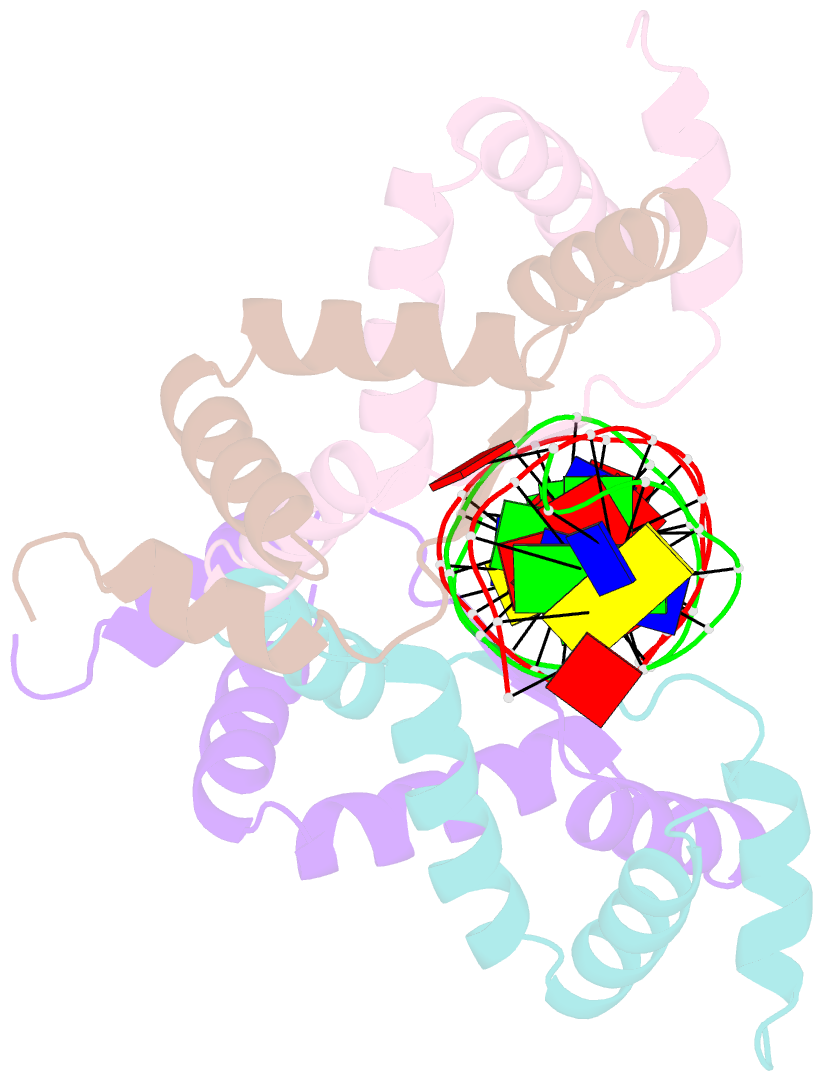
- Summary
- Structure of sulfolobus solfataricus segb-DNA complex
- Reference
- Yen CY, Lin MG, Chen BW, Ng IW, Read N, Kabli AF, Wu CT, Shen YY, Chen CH, Barilla D, Sun YJ, Hsiao CD (2021): "Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly." Nucleic Acids Res., 49, 13150-13164. doi: 10.1093/nar/gkab1155.
- Abstract
- Genome segregation is a vital process in all organisms. Chromosome partitioning remains obscure in Archaea, the third domain of life. Here, we investigated the SegAB system from Sulfolobus solfataricus. SegA is a ParA Walker-type ATPase and SegB is a site-specific DNA-binding protein. We determined the structures of both proteins and those of SegA-DNA and SegB-DNA complexes. The SegA structure revealed an atypical, novel non-sandwich dimer that binds DNA either in the presence or in the absence of ATP. The SegB structure disclosed a ribbon-helix-helix motif through which the protein binds DNA site specifically. The association of multiple interacting SegB dimers with the DNA results in a higher order chromatin-like structure. The unstructured SegB N-terminus plays an essential catalytic role in stimulating SegA ATPase activity and an architectural regulatory role in segrosome (SegA-SegB-DNA) formation. Electron microscopy results also provide a compact ring-like segrosome structure related to chromosome organization. These findings contribute a novel mechanistic perspective on archaeal chromosome segregation.