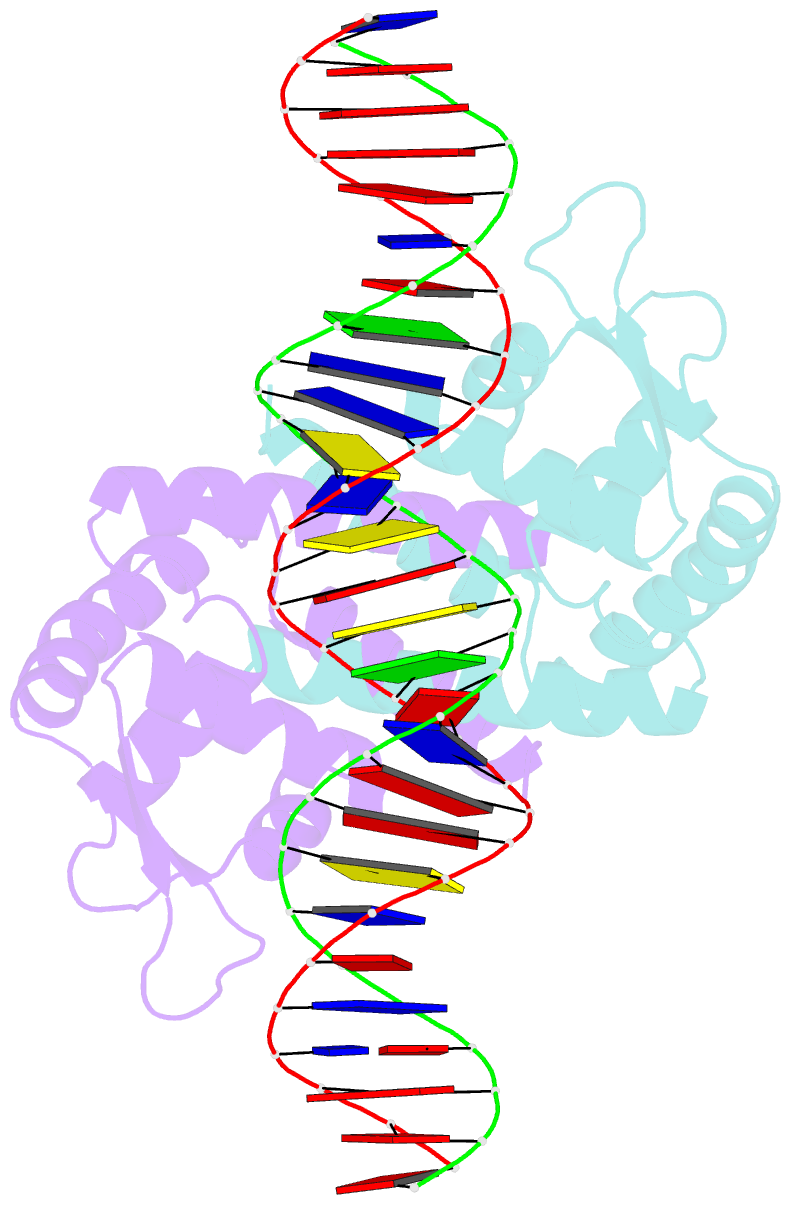
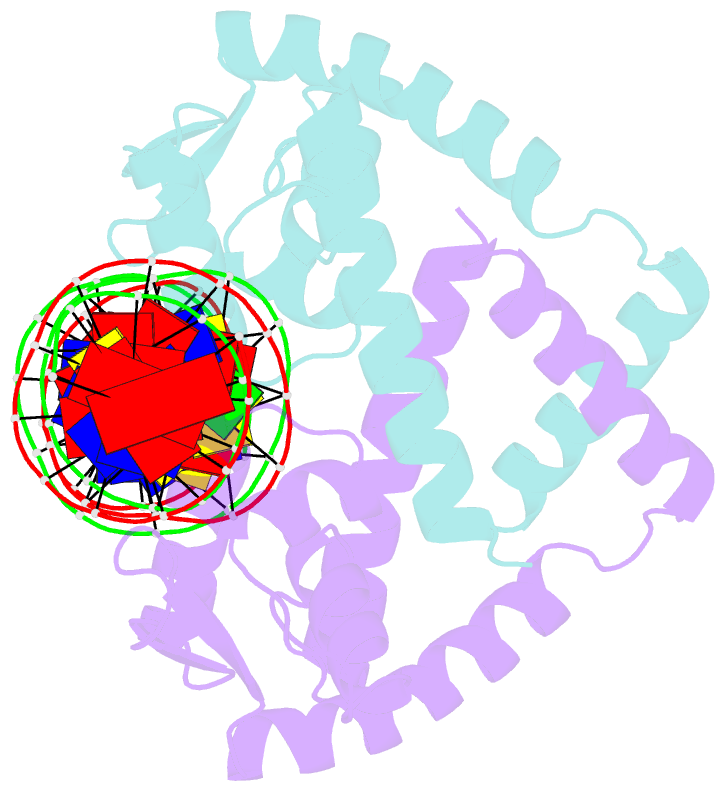
Summary information and primary citation
- PDB-id
- 7dvv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.49 Å)
- Summary
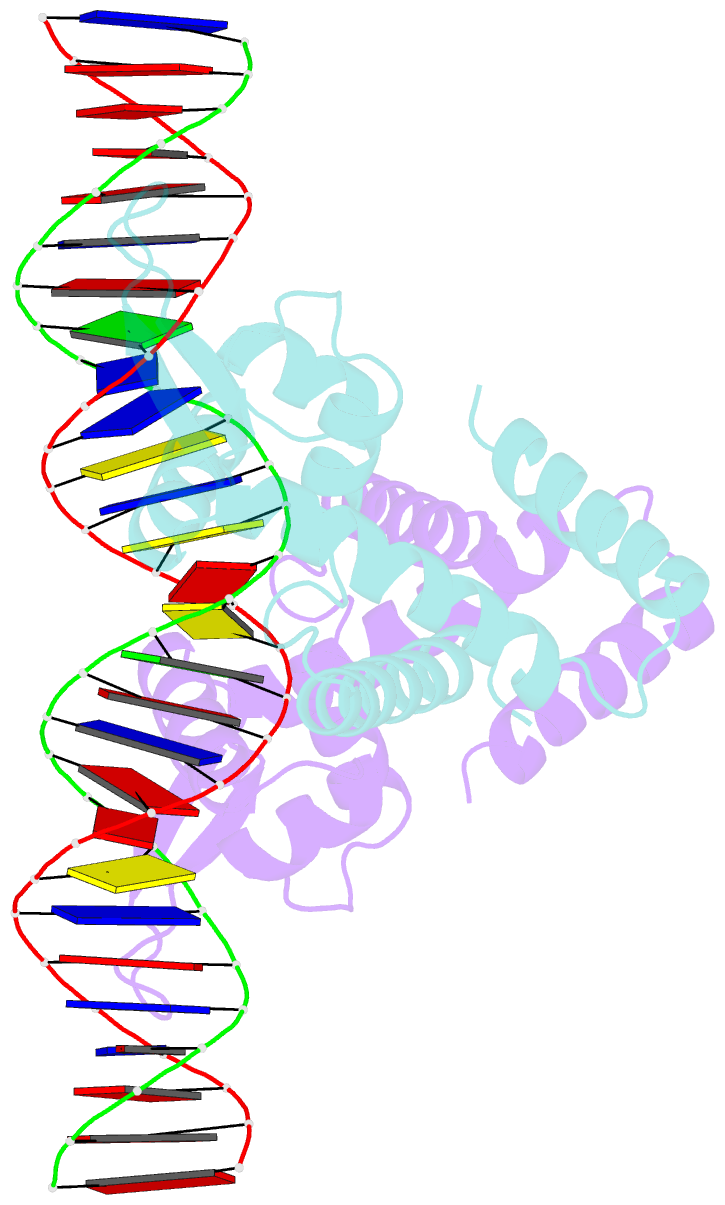
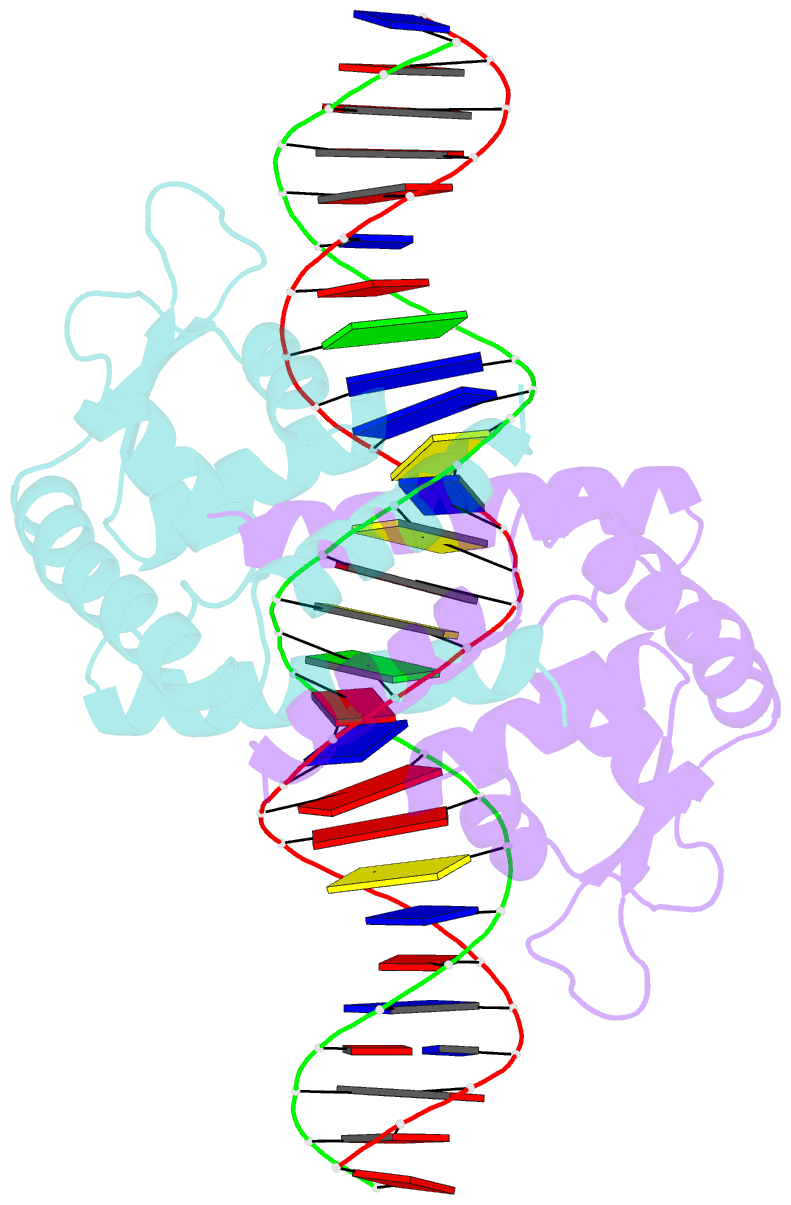
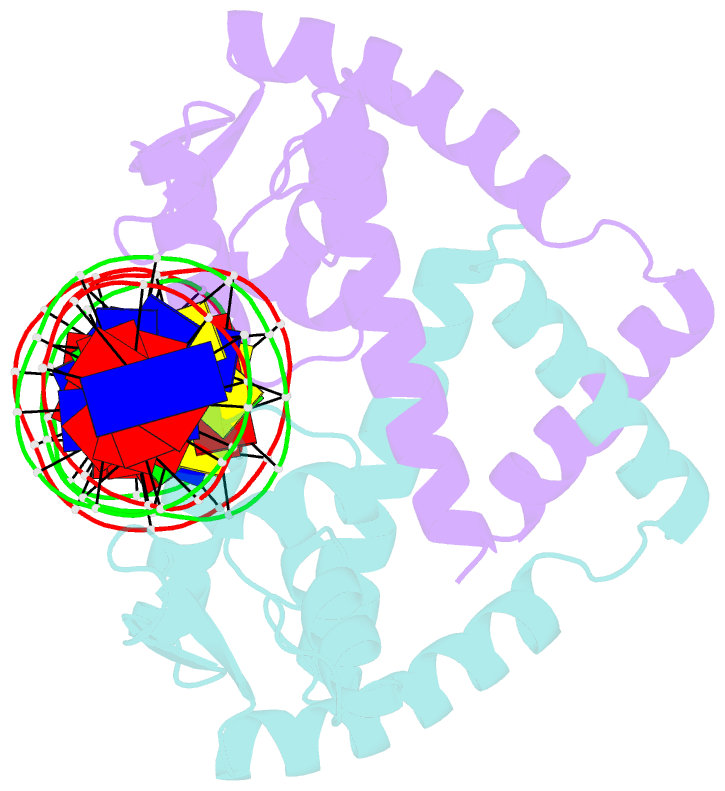
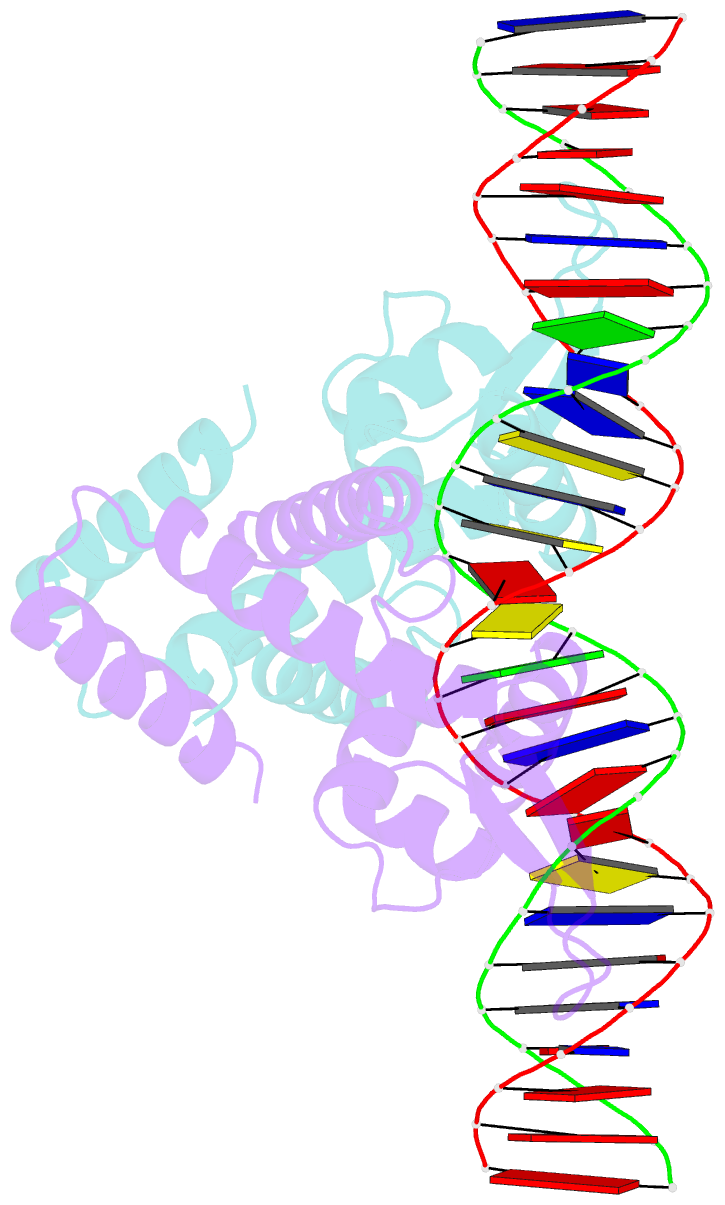
- Heme sensor protein pefr from streptococcus agalactiae bound to operator DNA (28-mer)
- Reference
- Nishinaga M, Sugimoto H, Nishitani Y, Nagai S, Nagatoishi S, Muraki N, Tosha T, Tsumoto K, Aono S, Shiro Y, Sawai H (2021): "Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival." Commun Biol, 4, 467. doi: 10.1038/s42003-021-01987-5.
- Abstract
- Hemes (iron-porphyrins) are critical for biological processes in all organisms. Hemolytic bacteria survive by acquiring b-type heme from hemoglobin in red blood cells from their animal hosts. These bacteria avoid the cytotoxicity of excess heme during hemolysis by expressing heme-responsive sensor proteins that act as transcriptional factors to regulate the heme efflux system in response to the cellular heme concentration. Here, the underlying regulatory mechanisms were investigated using crystallographic, spectroscopic, and biochemical studies to understand the structural basis of the heme-responsive sensor protein PefR from Streptococcus agalactiae, a causative agent of neonatal life-threatening infections. Structural comparison of heme-free PefR, its complex with a target DNA, and heme-bound PefR revealed that unique heme coordination controls a >20 Å structural rearrangement of the DNA binding domains to dissociate PefR from the target DNA. We also found heme-bound PefR stably binds exogenous ligands, including carbon monoxide, a by-product of the heme degradation reaction.