Summary information and primary citation
- PDB-id
- 7egq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- cryo-EM (3.35 Å)
- Summary
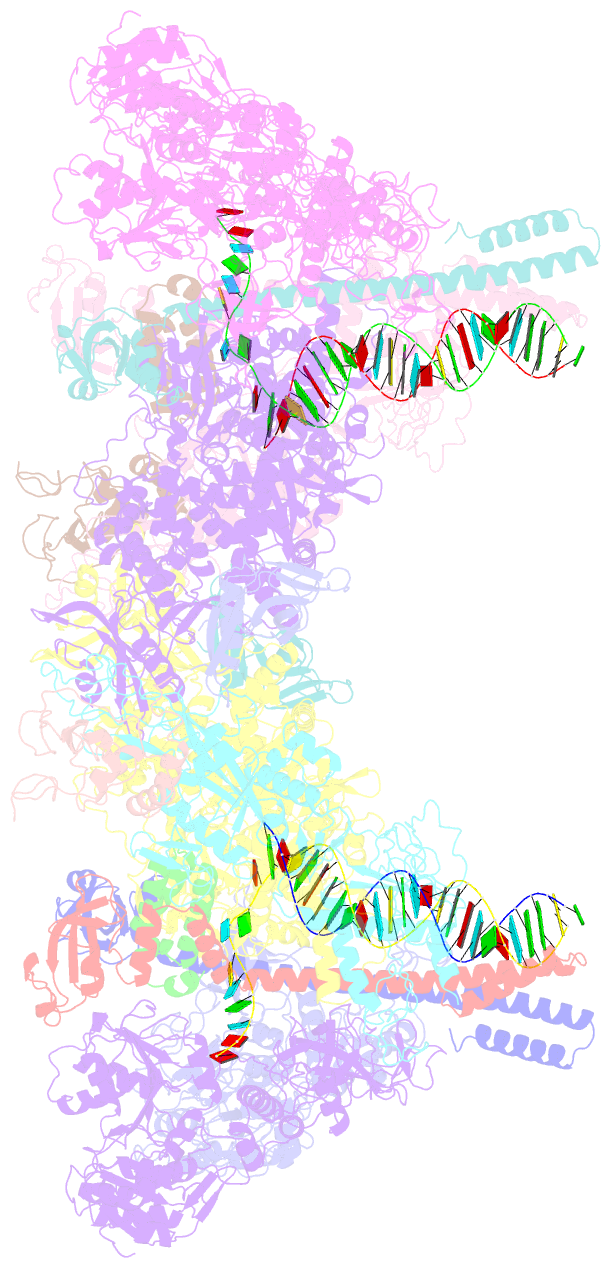
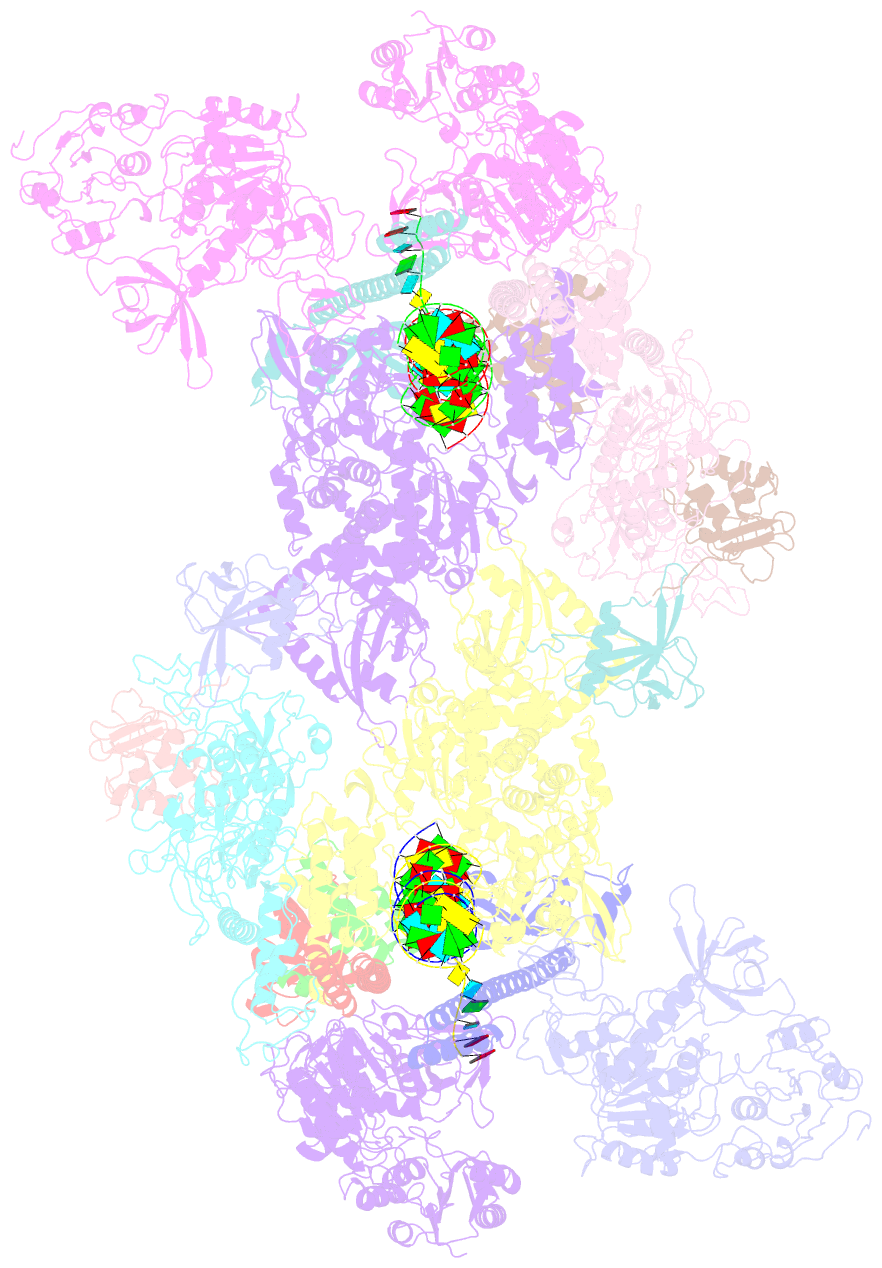
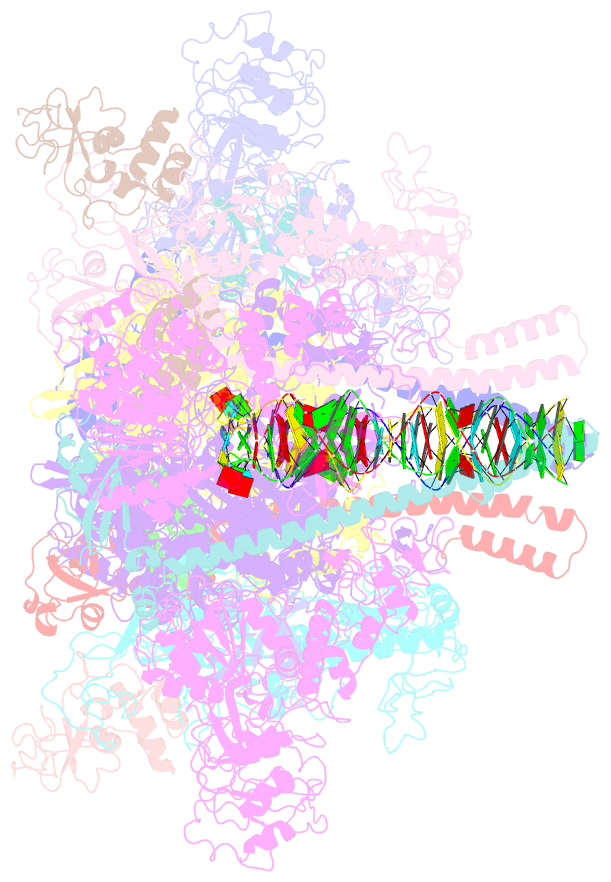
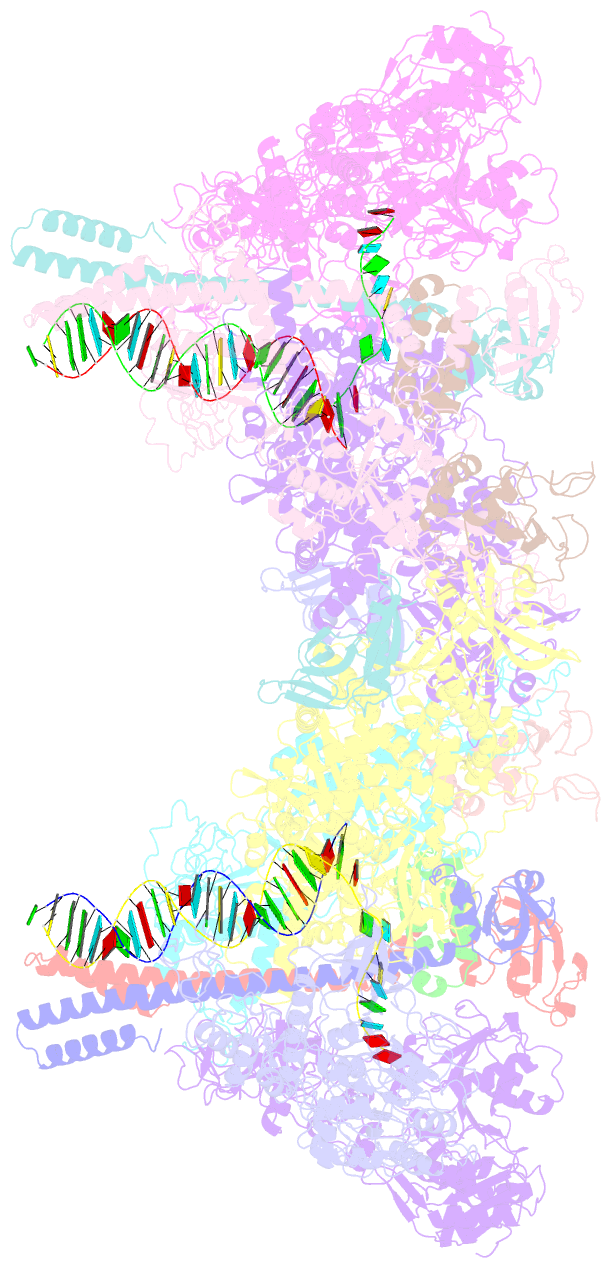
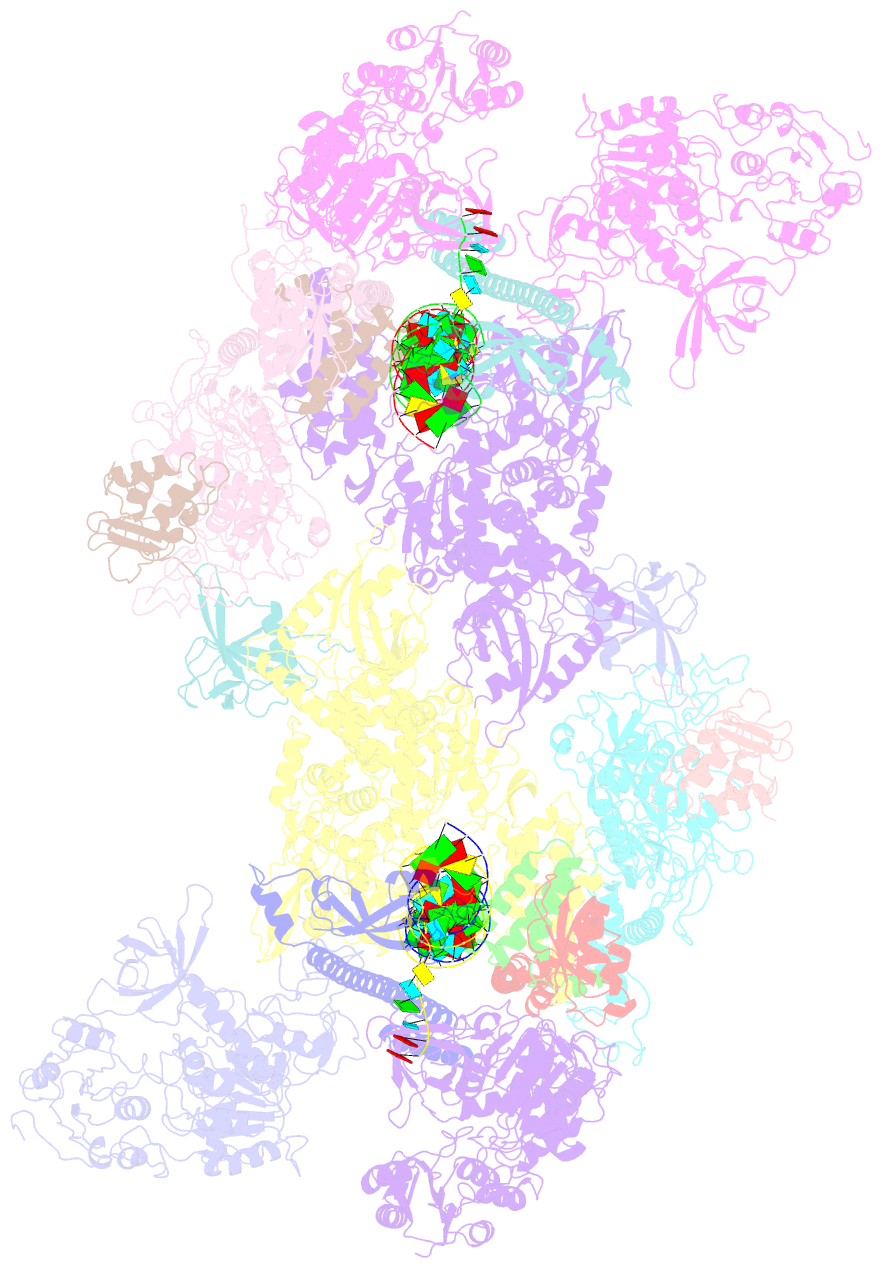
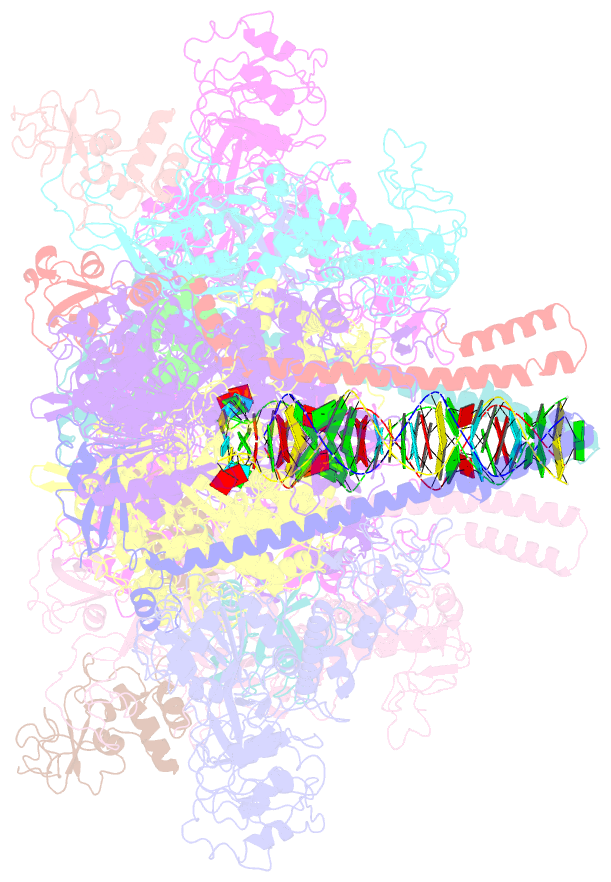
- Co-transcriptional capping machineries in sars-cov-2 rtc: coupling of n7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading
- Reference
- Yan L, Yang Y, Li M, Zhang Y, Zheng L, Ge J, Huang YC, Liu Z, Wang T, Gao S, Zhang R, Huang YY, Guddat LW, Gao Y, Rao Z, Lou Z (2021): "Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading." Cell, 184, 3474-3485.e11. doi: 10.1016/j.cell.2021.05.033.
- Abstract
- The capping of mRNA and the proofreading play essential roles in SARS-CoV-2 replication and transcription. Here, we present the cryo-EM structure of the SARS-CoV-2 replication-transcription complex (RTC) in a form identified as Cap(0)-RTC, which couples a co-transcriptional capping complex (CCC) composed of nsp12 NiRAN, nsp9, the bifunctional nsp14 possessing an N-terminal exoribonuclease (ExoN) and a C-terminal N7-methyltransferase (N7-MTase), and nsp10 as a cofactor of nsp14. Nsp9 and nsp12 NiRAN recruit nsp10/nsp14 into the Cap(0)-RTC, forming the N7-CCC to yield cap(0) (7MeGpppA) at 5' end of pre-mRNA. A dimeric form of Cap(0)-RTC observed by cryo-EM suggests an in trans backtracking mechanism for nsp14 ExoN to facilitate proofreading of the RNA in concert with polymerase nsp12. These results not only provide a structural basis for understanding co-transcriptional modification of SARS-CoV-2 mRNA but also shed light on how replication fidelity in SARS-CoV-2 is maintained.