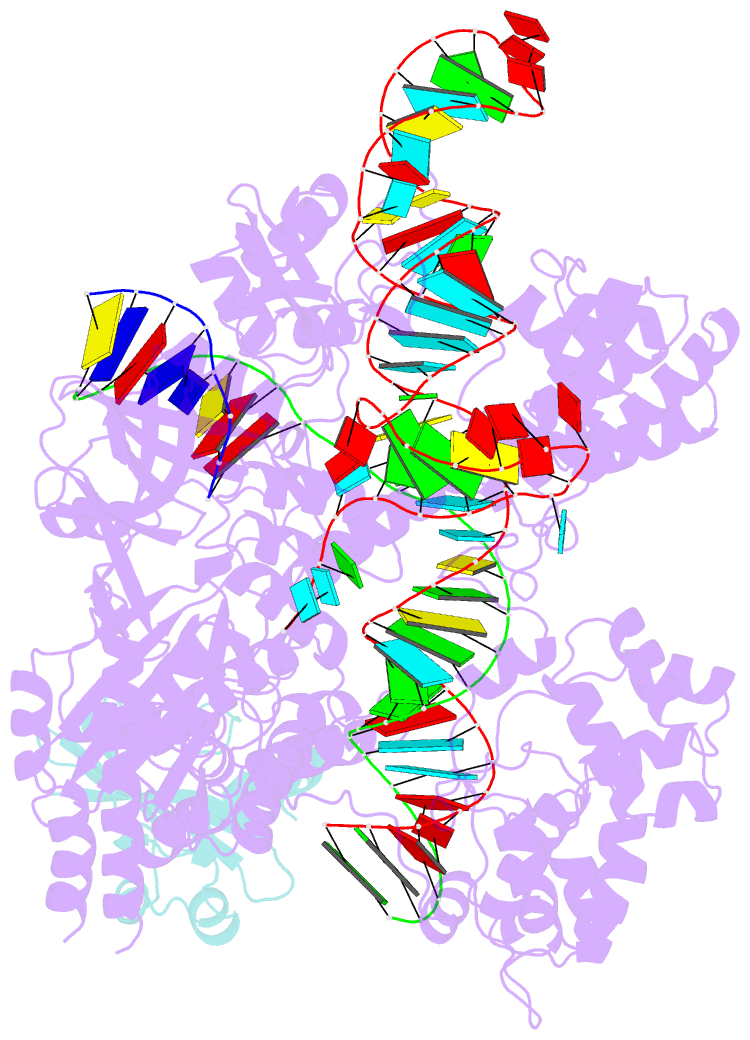
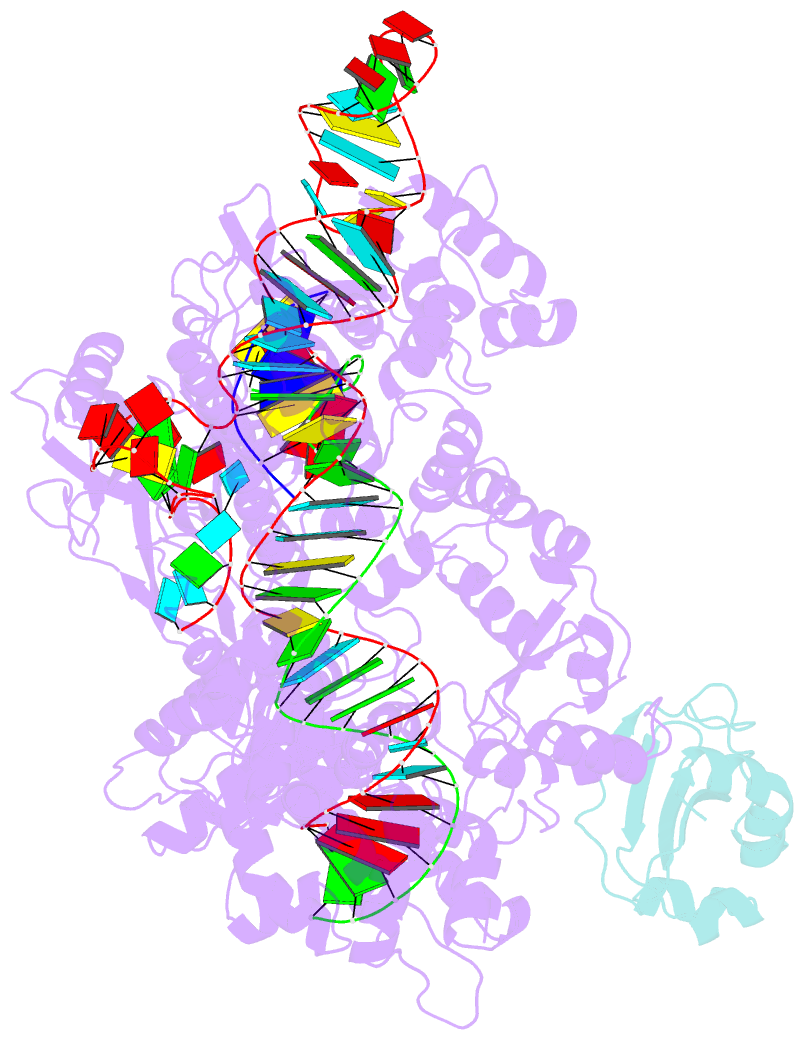
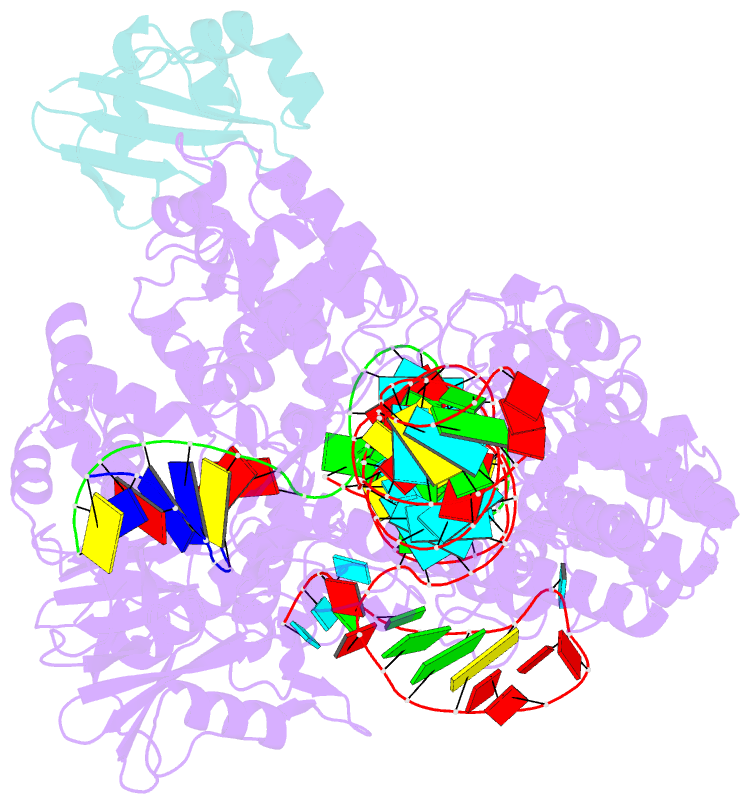
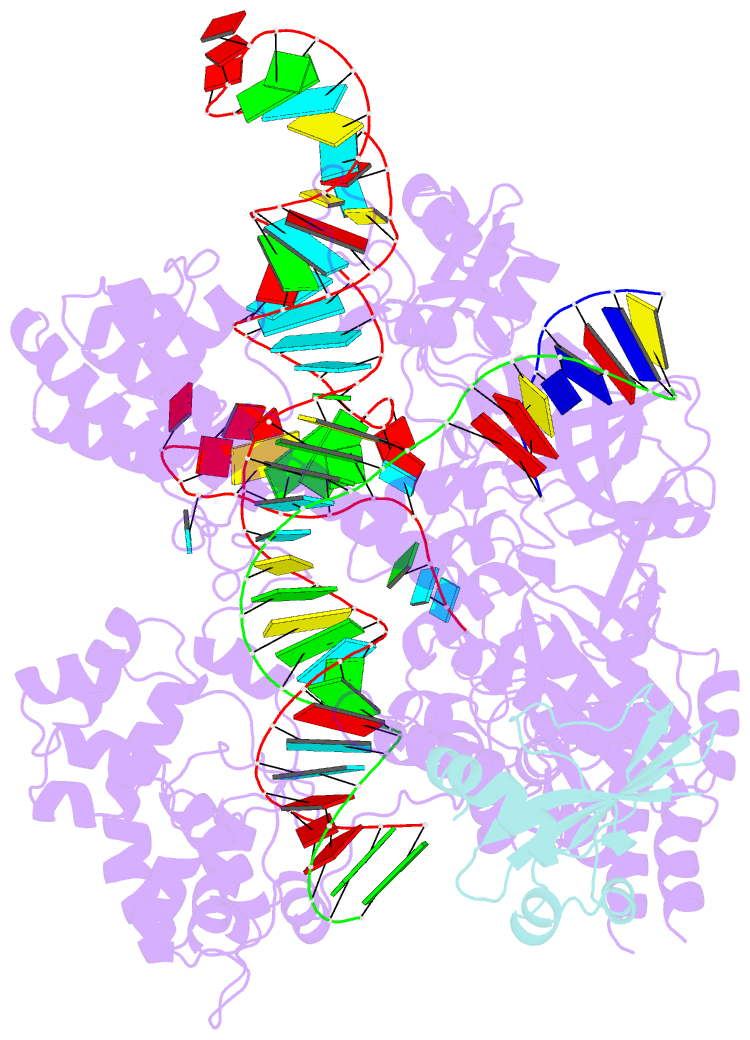
Summary information and primary citation
- PDB-id
- 7el1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- unknown function
- Method
- X-ray (2.22 Å)
- Summary
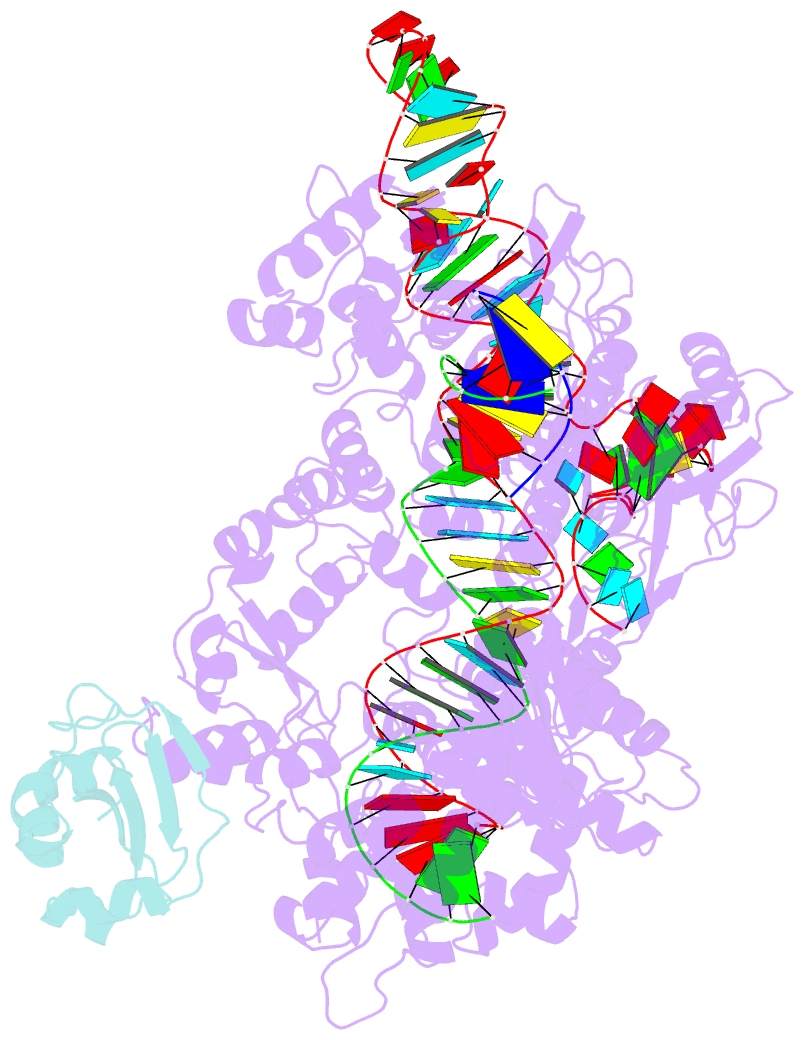
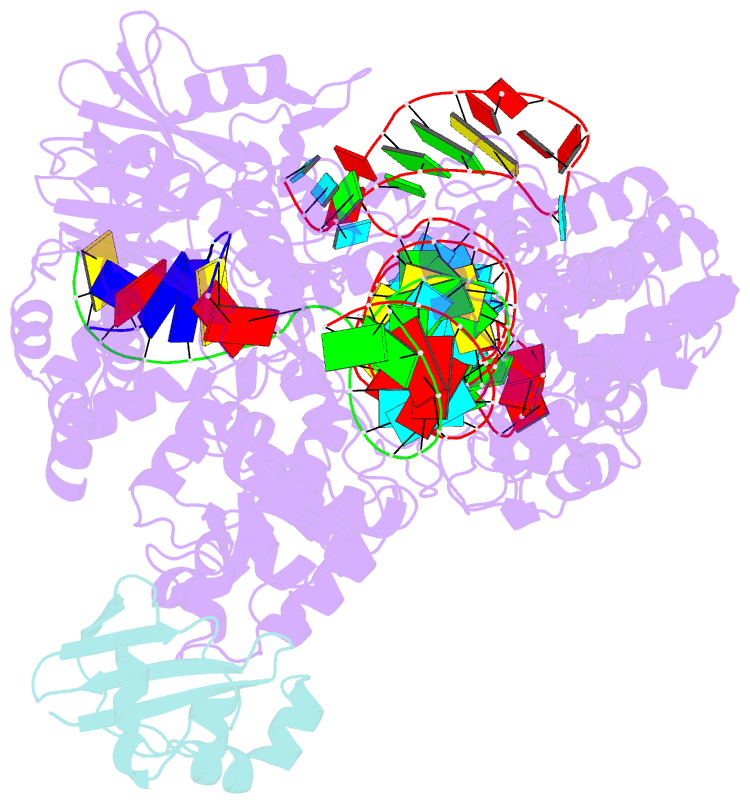
- Structure of a protein from bacteria
- Reference
- Liu H, Zhu Y, Lu Z, Huang Z (2021): "Structural basis of Staphylococcus aureus Cas9 inhibition by AcrIIA14." Nucleic Acids Res., 49, 6587-6595. doi: 10.1093/nar/gkab487.
- Abstract
- Bacteriophages have evolved a range of anti-CRISPR proteins (Acrs) to escape the adaptive immune system of prokaryotes, therefore Acrs can be used as switches to regulate gene editing. Herein, we report the crystal structure of a quaternary complex of AcrIIA14 bound SauCas9-sgRNA-dsDNA at 2.22 Å resolution, revealing the molecular basis for AcrIIA14 recognition and inhibition. Our structural and biochemical data analysis suggest that AcrIIA14 binds to a non-conserved region of SauCas9 HNH domain that is distinctly different from AcrIIC1 and AcrIIC3, with no significant effect on sgRNA or dsDNA binding. Further, our structural data shows that the allostery of the HNH domain close to the substrate DNA is sterically prevented by AcrIIA14 binding. In addition, the binding of AcrIIA14 triggers the conformational allostery of the HNH domain and the L1 linker within the SauCas9, driving them to make new interactions with the target-guide heteroduplex, enhancing the inhibitory ability of AcrIIA14. Our research both expands the current understanding of anti-CRISPRs and provides additional culues for the rational use of the CRISPR-Cas system in genome editing and gene regulation.