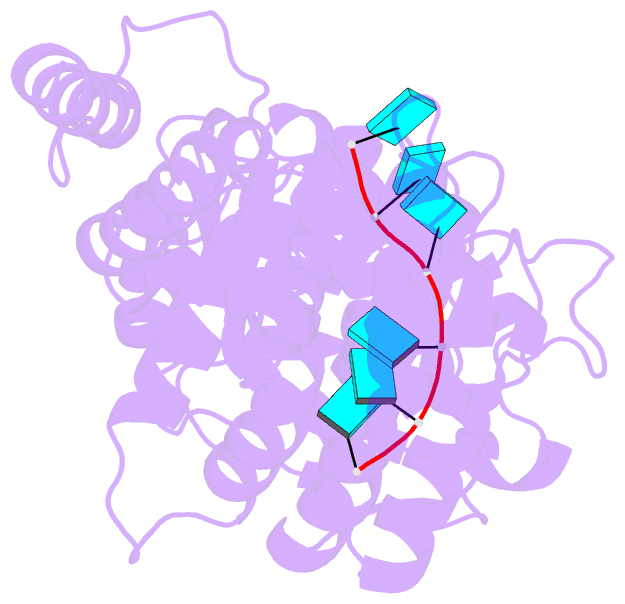
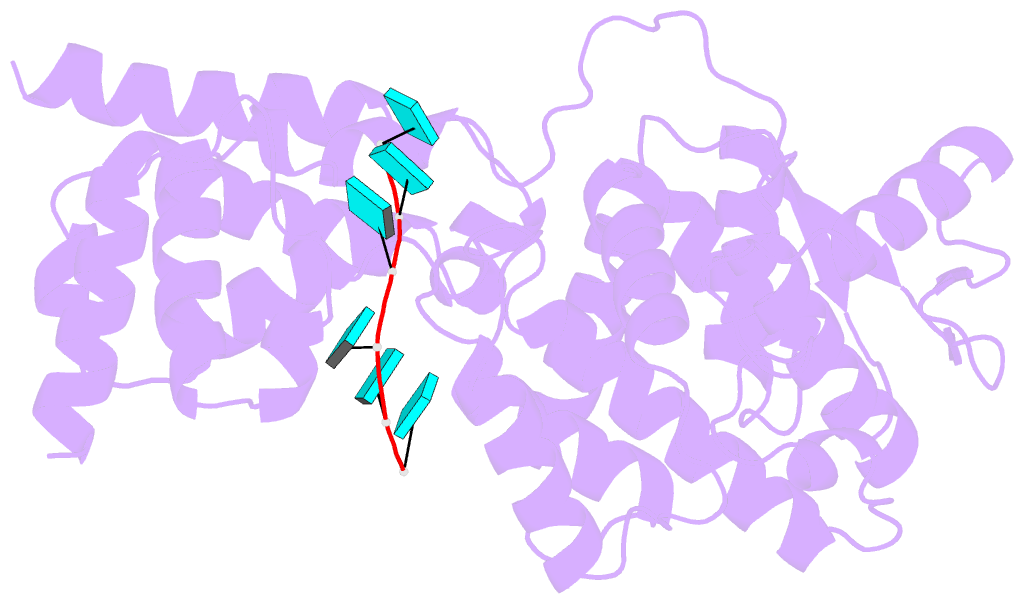
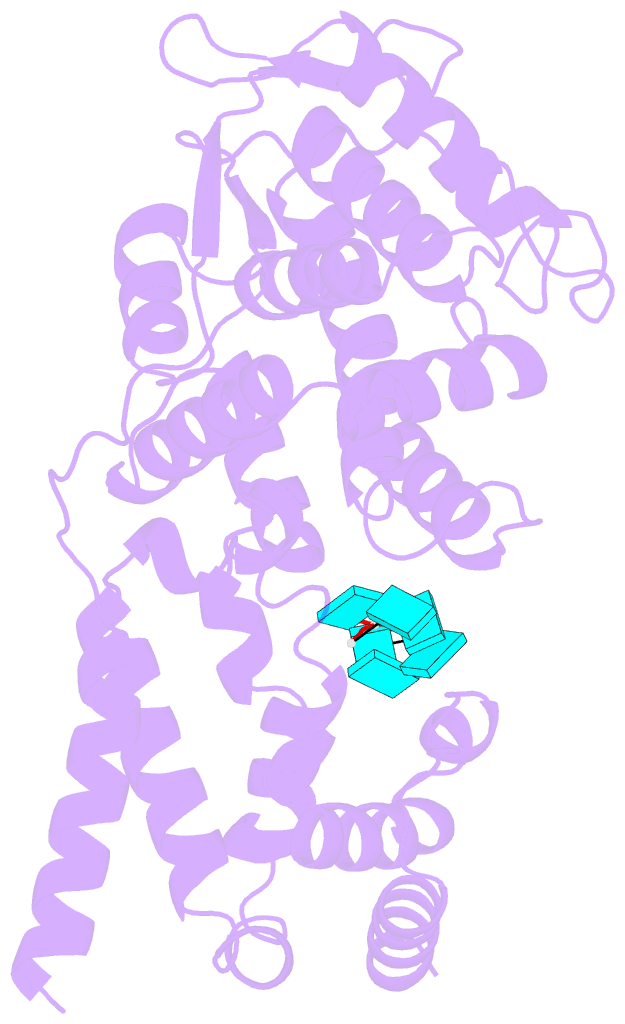
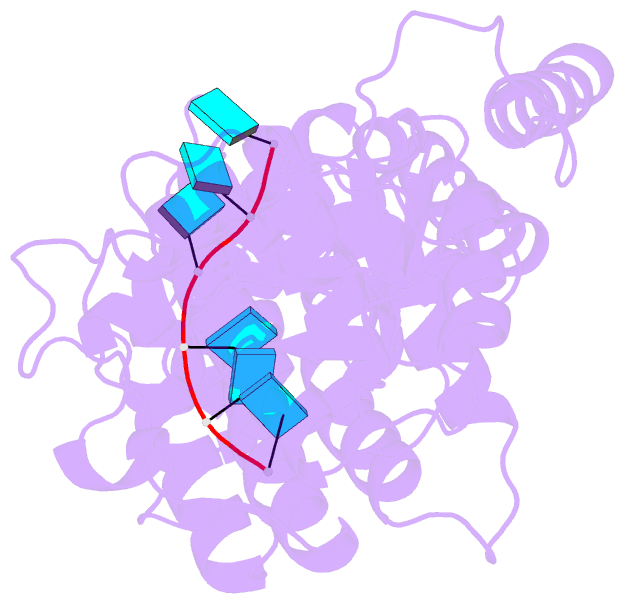
Summary information and primary citation
- PDB-id
- 7exa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (2.9 Å)
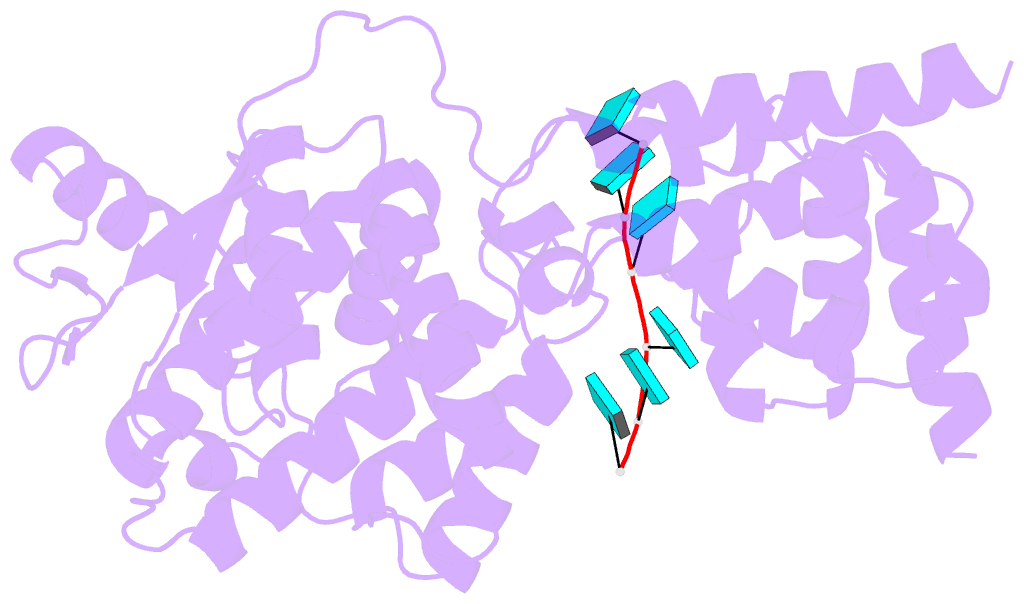
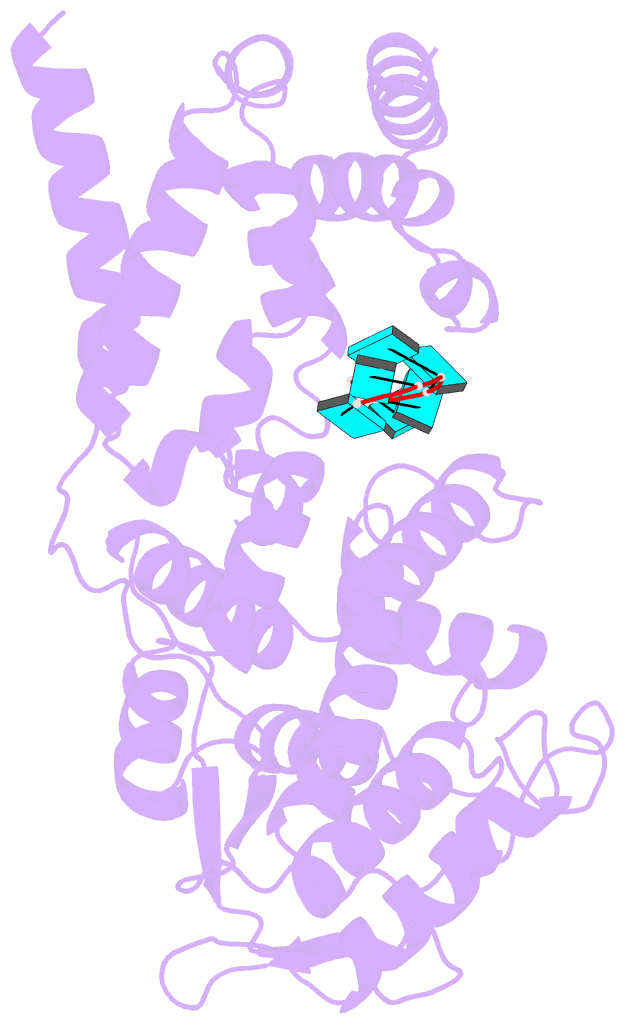
- Summary
- Structure of mumps virus nucleoprotein without c-arm
- Reference
- Shan H, Su X, Li T, Qin Y, Zhang N, Yang L, Ma L, Bai Y, Qi L, Liu Y, Shen QT (2021): "Structural plasticity of mumps virus nucleocapsids with cryo-EM structures." Commun Biol, 4, 833. doi: 10.1038/s42003-021-02362-0.
- Abstract
- Mumps virus (MuV) is a highly contagious human pathogen and frequently causes worldwide outbreaks despite available vaccines. Similar to other mononegaviruses such as Ebola and rabies, MuV uses a single-stranded negative-sense RNA as its genome, which is enwrapped by viral nucleoproteins into the helical nucleocapsid. The nucleocapsid acts as a scaffold for genome condensation and as a template for RNA replication and transcription. Conformational changes in the MuV nucleocapsid are required to switch between different activities, but the underlying mechanism remains elusive due to the absence of high-resolution structures. Here, we report two MuV nucleoprotein-RNA rings with 13 and 14 protomers, one stacked-ring filament and two nucleocapsids with distinct helical pitches, in dense and hyperdense states, at near-atomic resolutions using cryo-electron microscopy. Structural analysis of these in vitro assemblies indicates that the C-terminal tail of MuV nucleoprotein likely regulates the assembly of helical nucleocapsids, and the C-terminal arm may be relevant for the transition between the dense and hyperdense states of helical nucleocapsids. Our results provide the molecular mechanism for structural plasticity among different MuV nucleocapsids and create a possible link between structural plasticity and genome condensation.