Summary information and primary citation
- PDB-id
- 7f0r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (5.8 Å)
- Summary
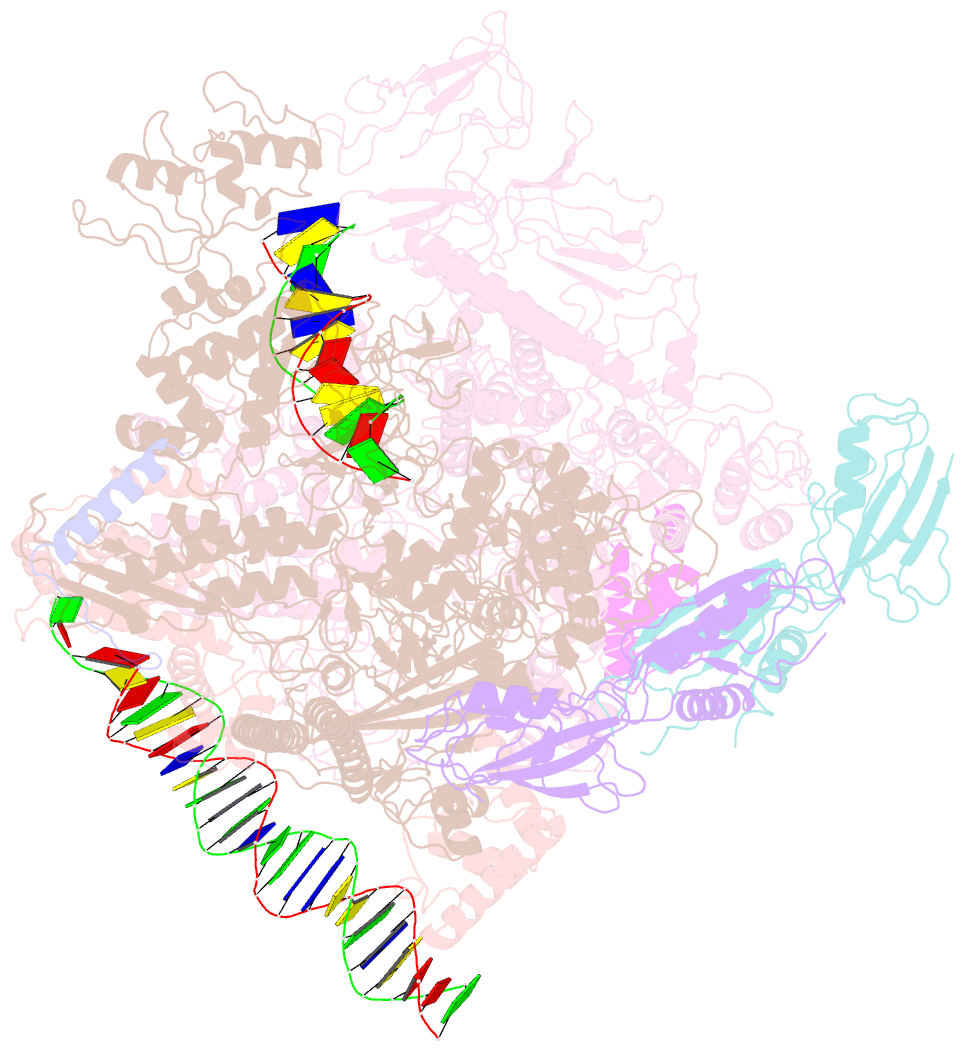
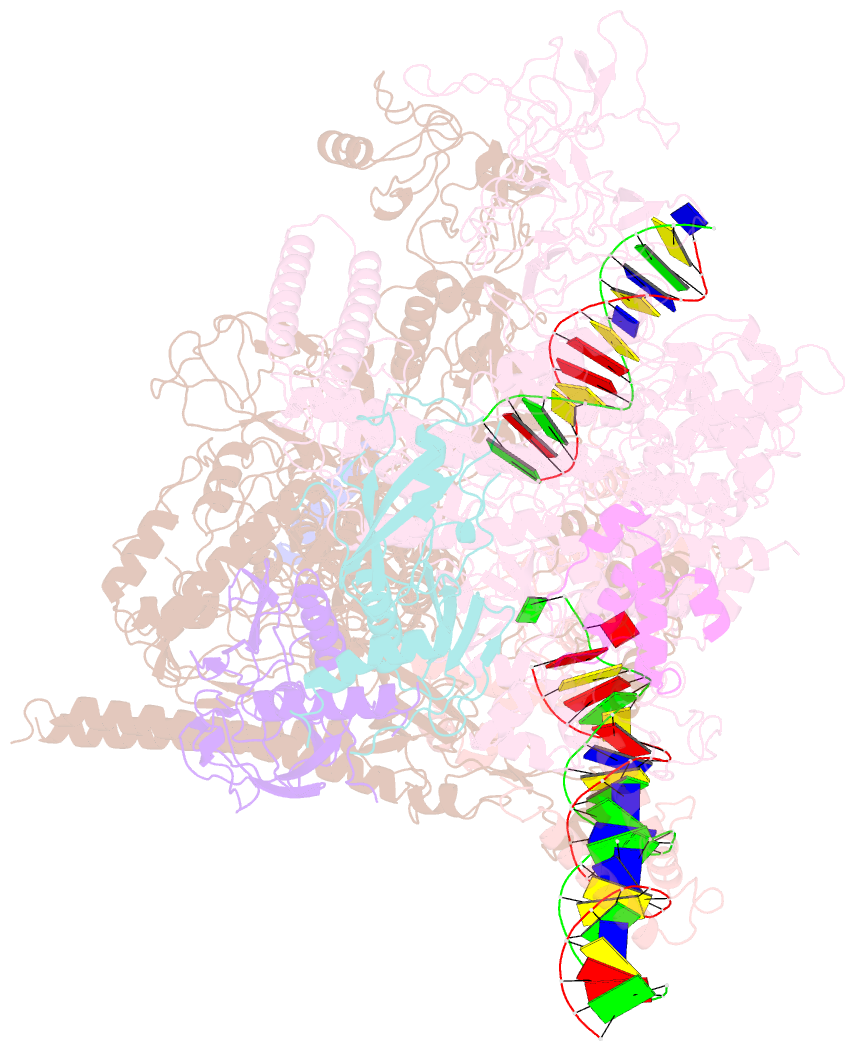
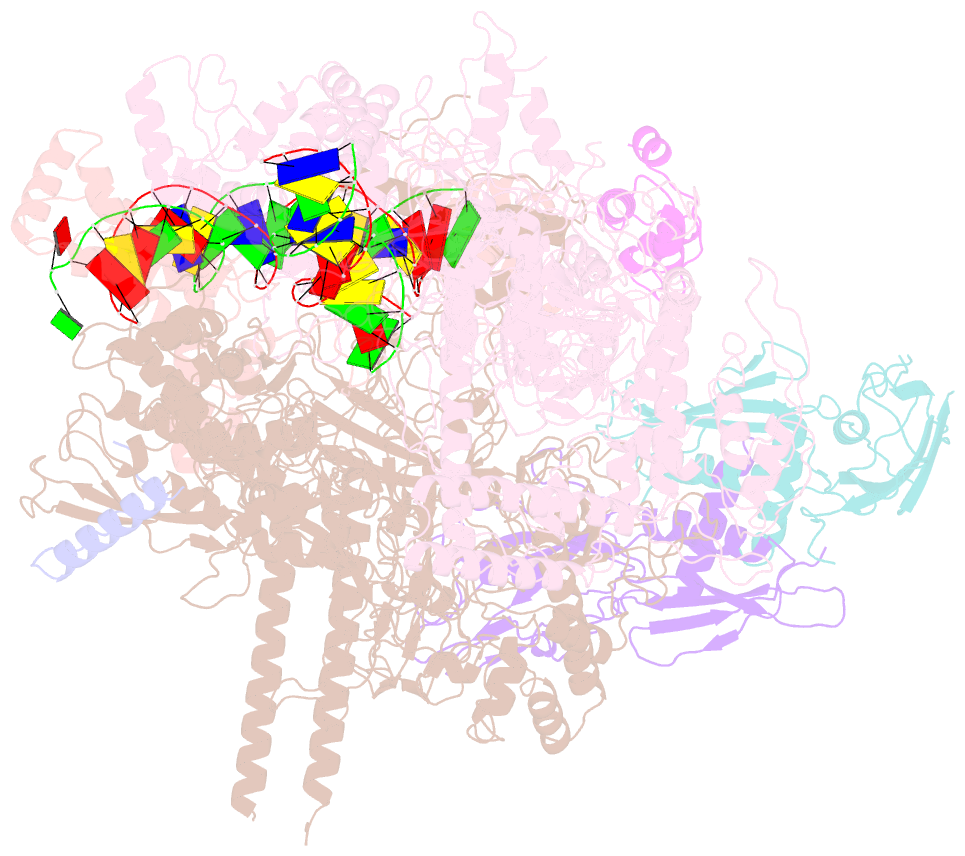
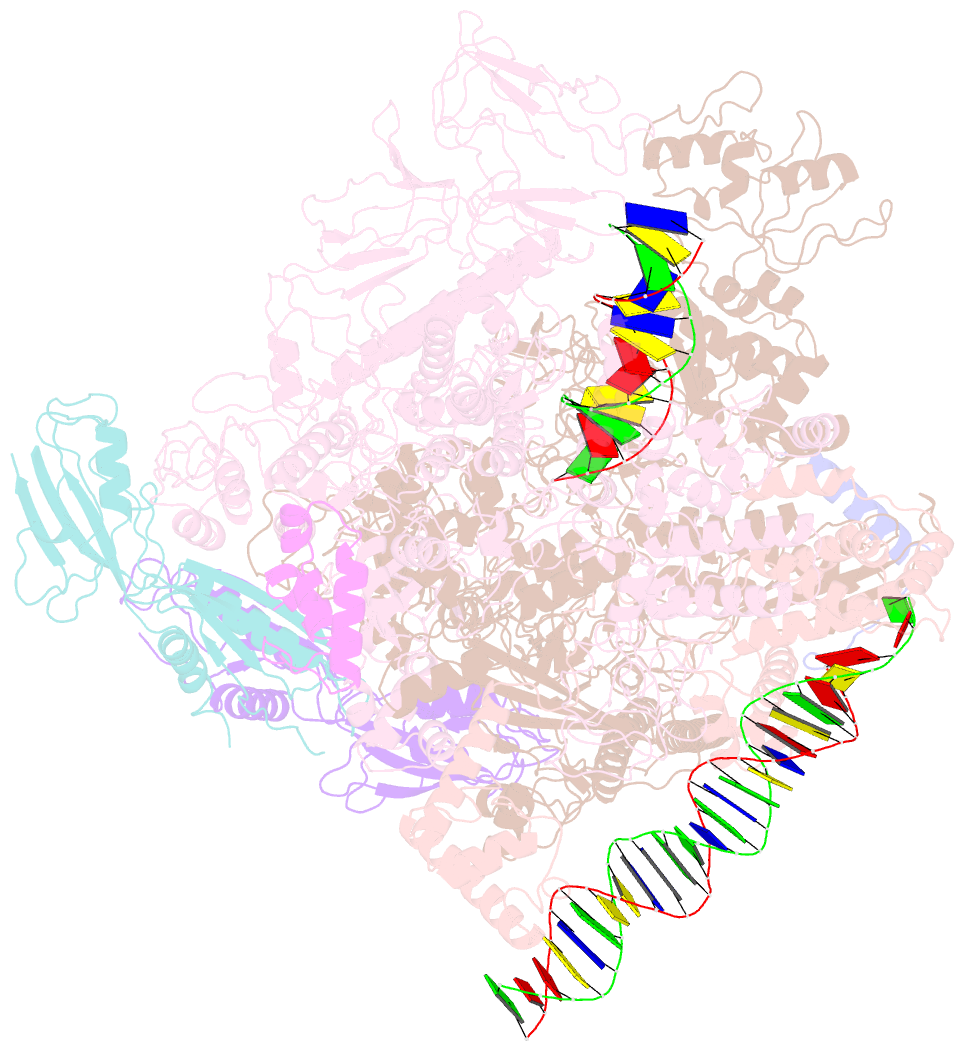
- cryo-EM structure of pseudomonas aeruginosa suta transcription activation complex
- Reference
- He D, You L, Wu X, Shi J, Wen A, Yan Z, Mu W, Fang C, Feng Y, Zhang Y (2022): "Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding." Nat Commun, 13, 4204. doi: 10.1038/s41467-022-31871-7.
- Abstract
- Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme comprising the stress-responsive σ factor σS (RNAP-σS). SutA shows no homology to previously characterized RNAP-binding proteins. The structure and mode of action of SutA remain unclear. Here we determined cryo-EM structures of Pae RNAP-σS holoenzyme, Pae RNAP-σS holoenzyme complexed with SutA, and Pae RNAP-σS transcription initiation complex comprising SutA. The structures show SutA pinches RNAP-β protrusion and facilitates promoter unwinding by wedging RNAP-β lobe open. Our results demonstrate that SutA clears an energetic barrier to facilitate promoter unwinding of RNAP-σS holoenzyme.