Summary information and primary citation
- PDB-id
- 7f1m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.1 Å)
- Summary
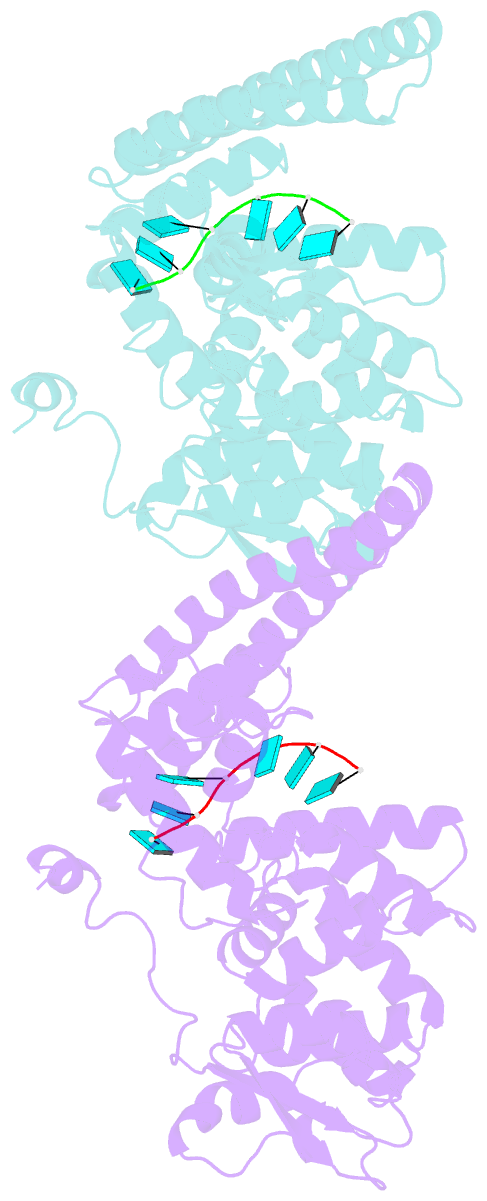
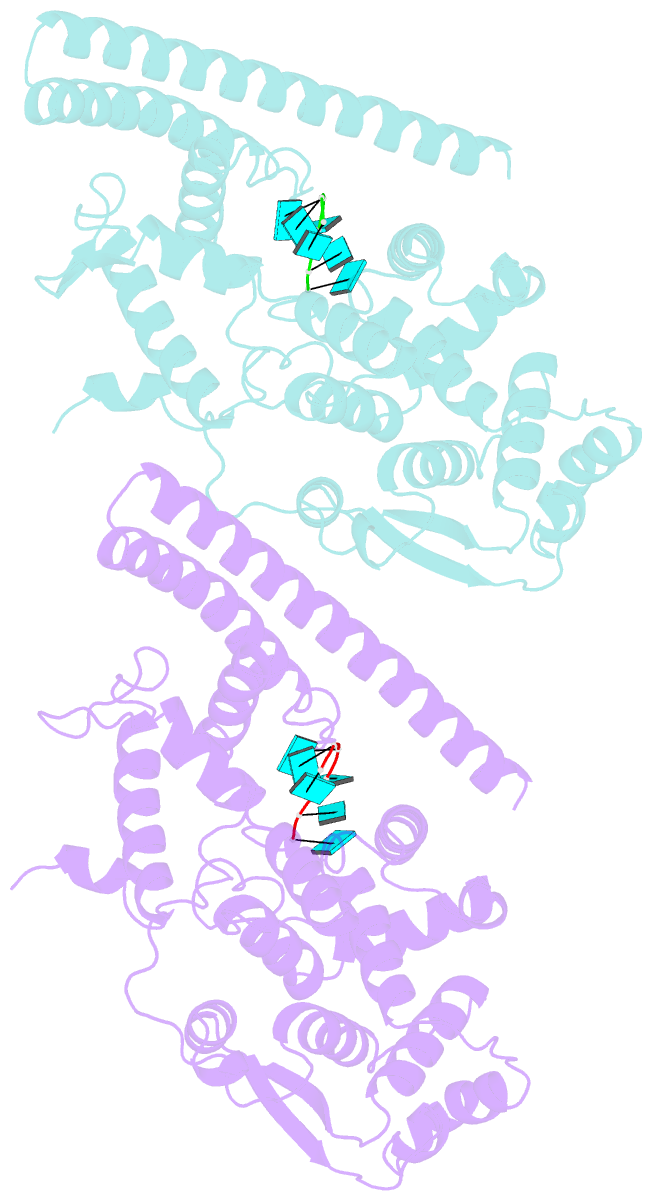
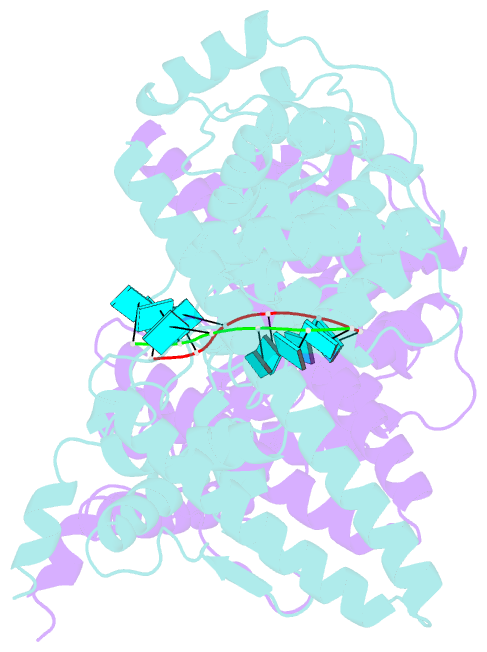
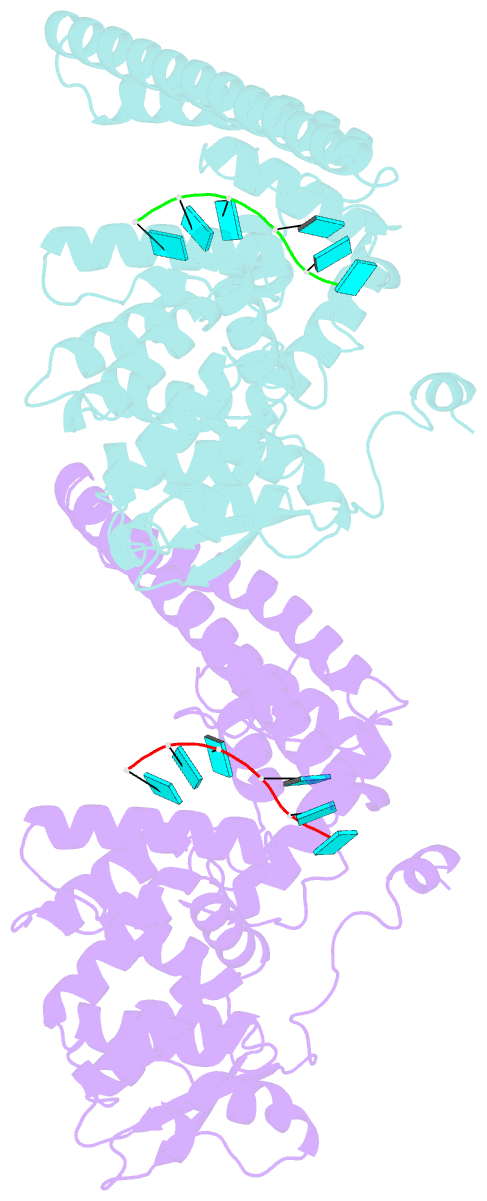
- Marburg virus nucleoprotein-RNA complex
- Reference
- Fujita-Fujiharu Y, Sugita Y, Takamatsu Y, Houri K, Igarashi M, Muramoto Y, Nakano M, Tsunoda Y, Taniguchi I, Becker S, Noda T (2022): "Structural insight into Marburg virus nucleoprotein-RNA complex formation." Nat Commun, 13, 1191. doi: 10.1038/s41467-022-28802-x.
- Abstract
- The nucleoprotein (NP) of Marburg virus (MARV), a close relative of Ebola virus (EBOV), encapsidates the single-stranded, negative-sense viral genomic RNA (vRNA) to form the helical NP-RNA complex. The NP-RNA complex constitutes the core structure for the assembly of the nucleocapsid that is responsible for viral RNA synthesis. Although appropriate interactions among NPs and RNA are required for the formation of nucleocapsid, the structural basis of the helical assembly remains largely elusive. Here, we show the structure of the MARV NP-RNA complex determined using cryo-electron microscopy at a resolution of 3.1 Å. The structures of the asymmetric unit, a complex of an NP and six RNA nucleotides, was very similar to that of EBOV, suggesting that both viruses share common mechanisms for the nucleocapsid formation. Structure-based mutational analysis of both MARV and EBOV NPs identified key residues for helical assembly and subsequent viral RNA synthesis. Importantly, most of the residues identified were conserved in both viruses. These findings provide a structural basis for understanding the nucleocapsid formation and contribute to the development of novel antivirals against MARV and EBOV.