Summary information and primary citation
- PDB-id
- 7f9h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (1.78 Å)
- Summary
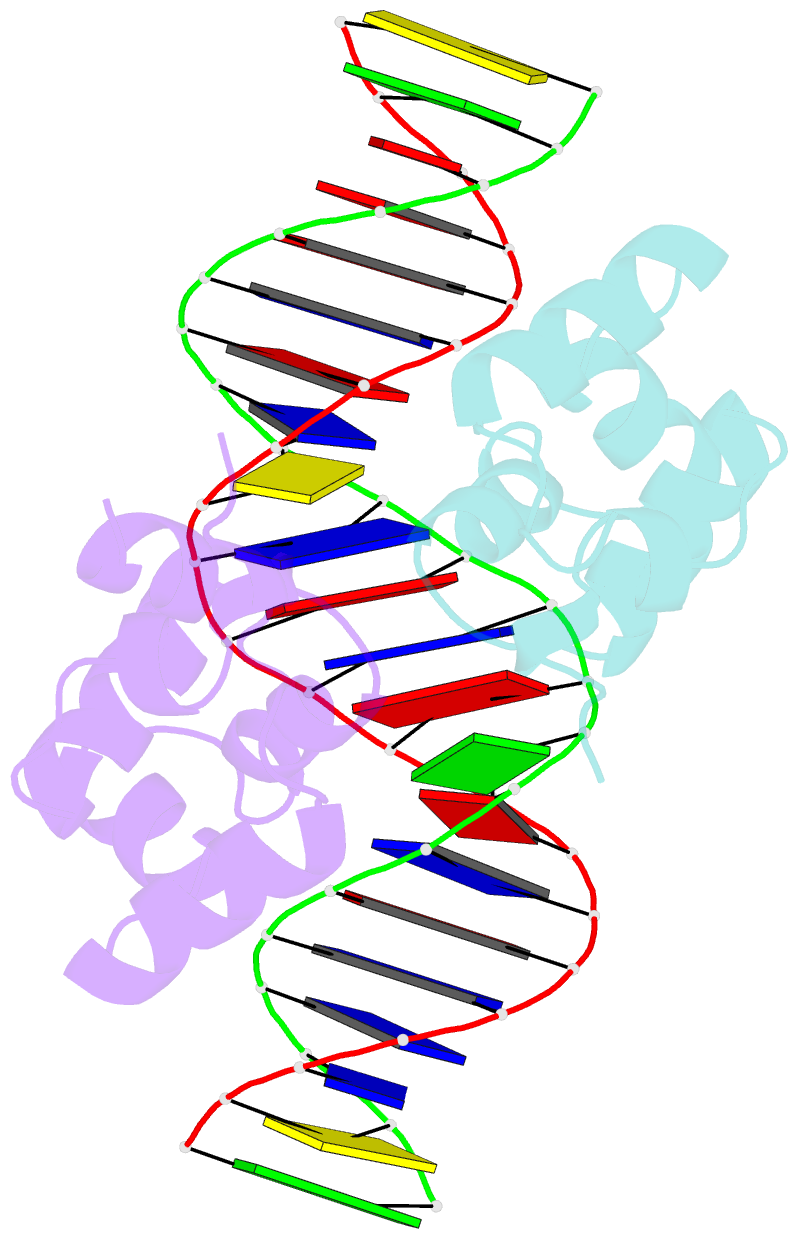
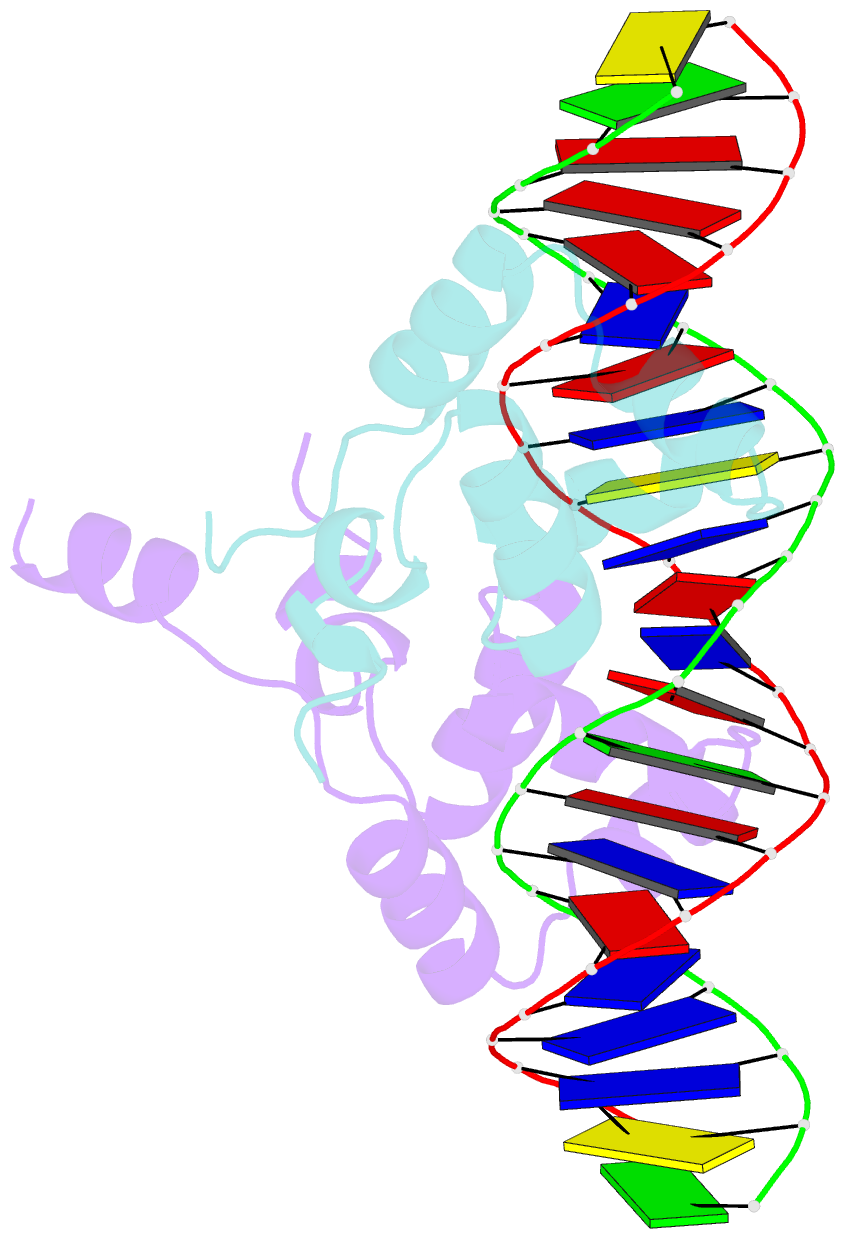
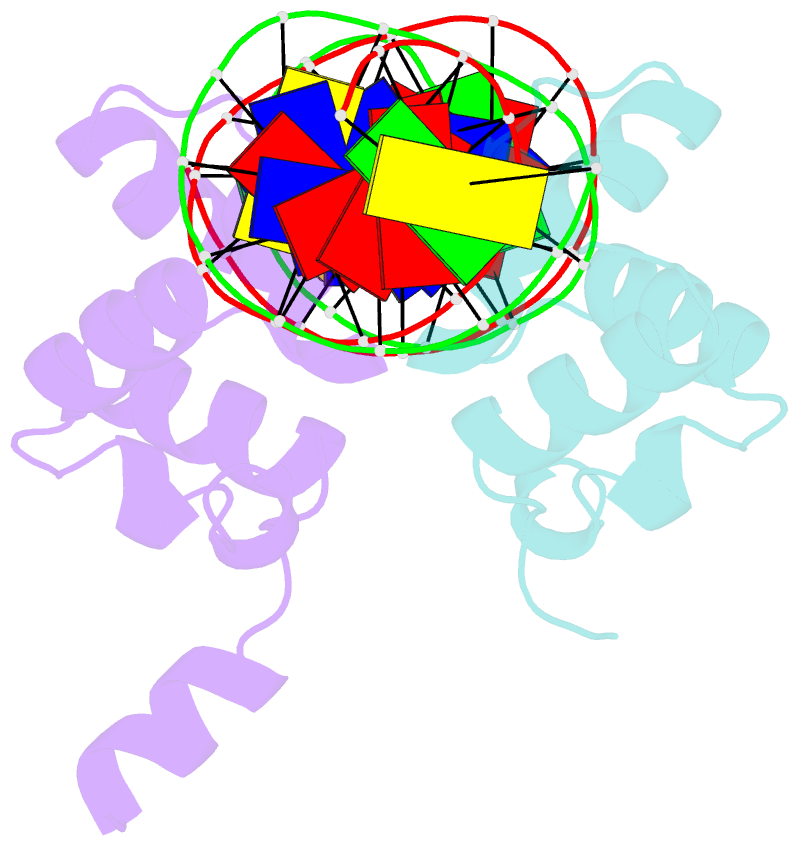
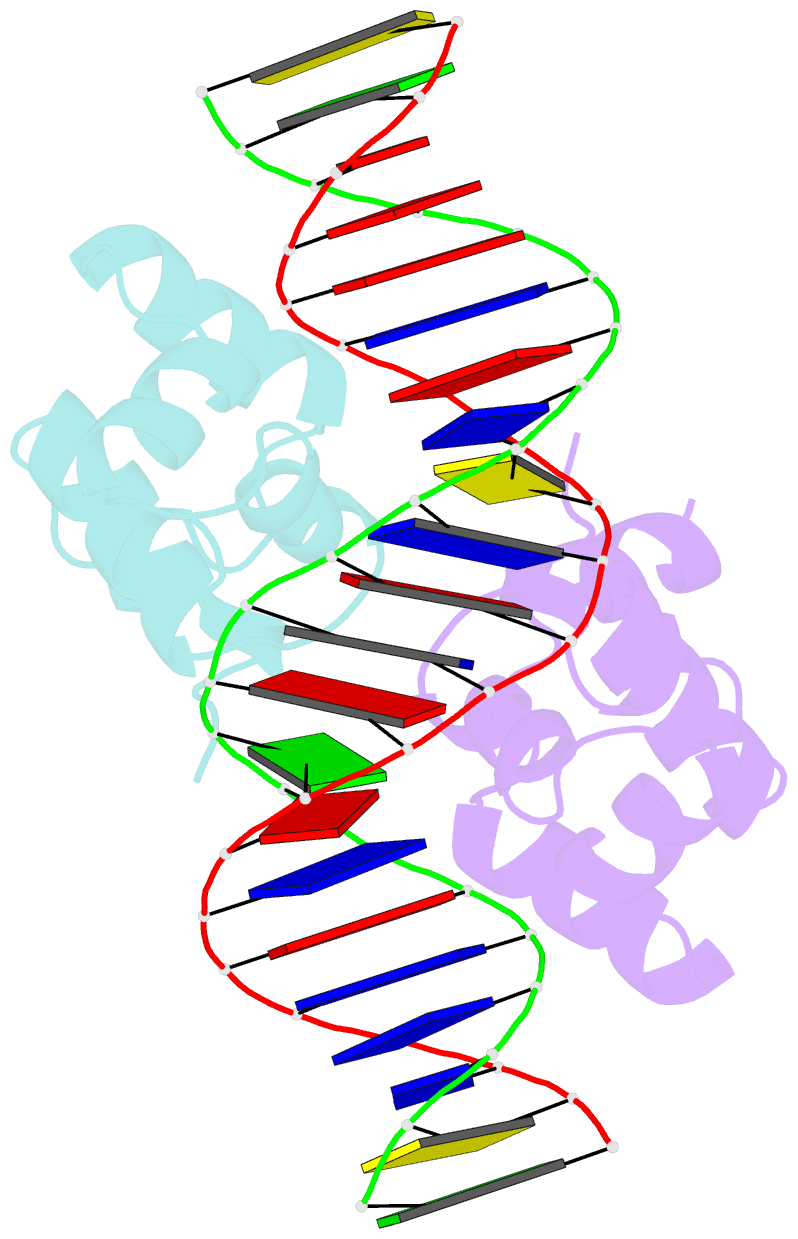
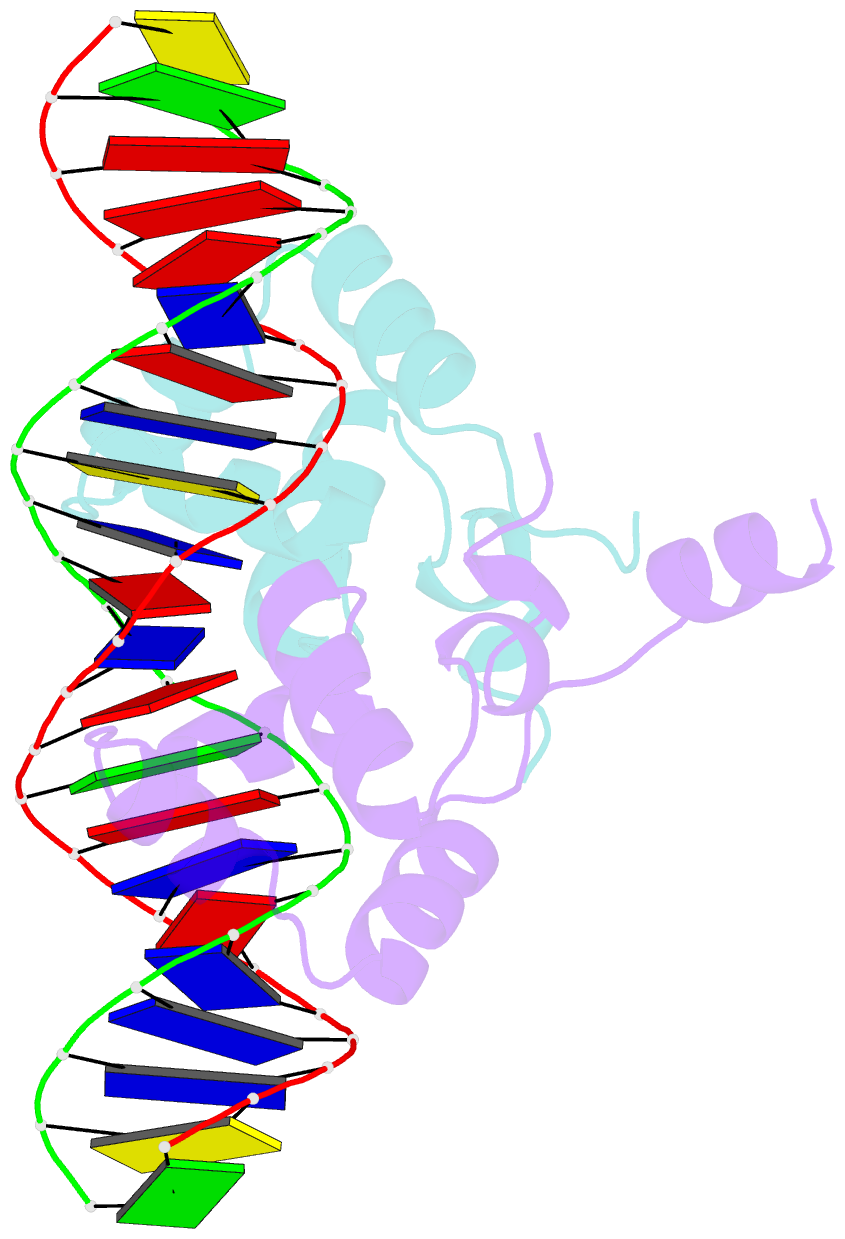
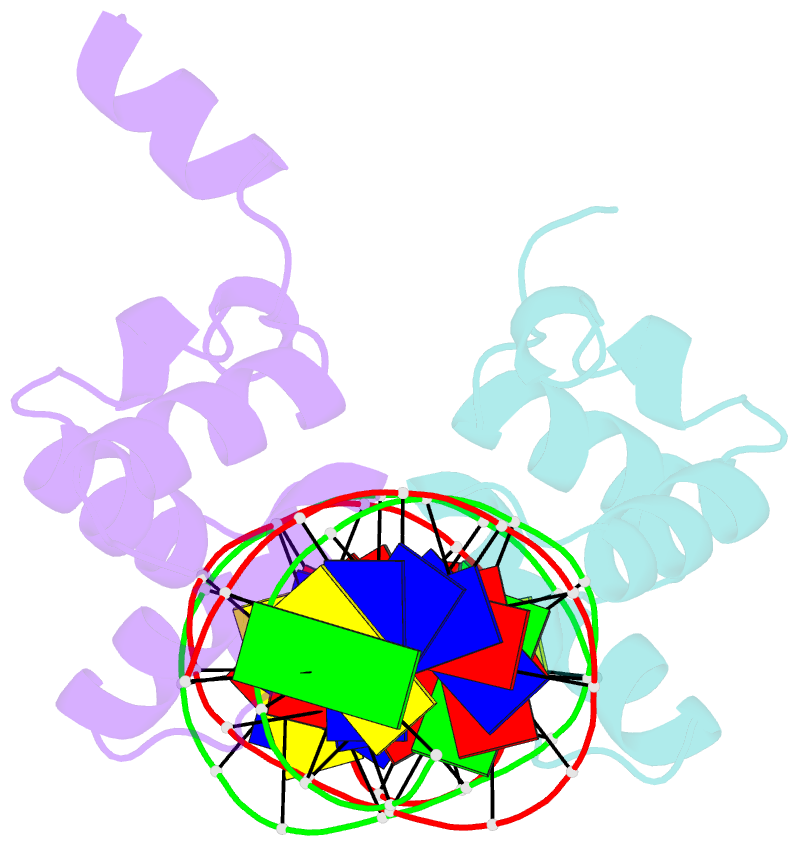
- Complex structure of enrr-DNA
- Reference
- Ma R, Liu Y, Gan J, Qiao H, Ma J, Zhang Y, Bu Y, Shao S, Zhang Y, Wang Q (2022): "Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding." Nucleic Acids Res., 50, 3777-3798. doi: 10.1093/nar/gkac180.
- Abstract
- Type III and type VI secretion systems (T3/T6SS) are encoded in horizontally acquired genomic islands (GIs) that play crucial roles in evolution and virulence in bacterial pathogens. T3/T6SS expression is subjected to tight control by the host xenogeneic silencer H-NS, but how this mechanism is counteracted remains to be illuminated. Here, we report that xenogeneic nucleoid-associated protein EnrR encoded in a GI is essential for virulence in pathogenic bacteria Edwardsiella and Salmonella. We showed that EnrR plays critical roles in T3/T6SS expression in these bacteria. Various biochemical and genetic analyses demonstrated that EnrR binds and derepresses the promoter of esrB, the critical regulator of T3/T6SS, to promote their expression by competing with H-NS. Additionally, EnrR targets AT-rich regions, globally modulates the expression of ∼363 genes and is involved in various cellular processes. Crystal structures of EnrR in complex with a specific AT-rich palindromic DNA revealed a new DNA-binding mode that involves conserved HTH-mediated interactions with the major groove and contacts of its N-terminal extension to the minor groove in the symmetry-related duplex. Collectively, these data demonstrate that EnrR is a virulence activator that can antagonize H-NS, highlighting a unique mechanism by which bacterial xenogeneic regulators recognize and regulate foreign DNA.