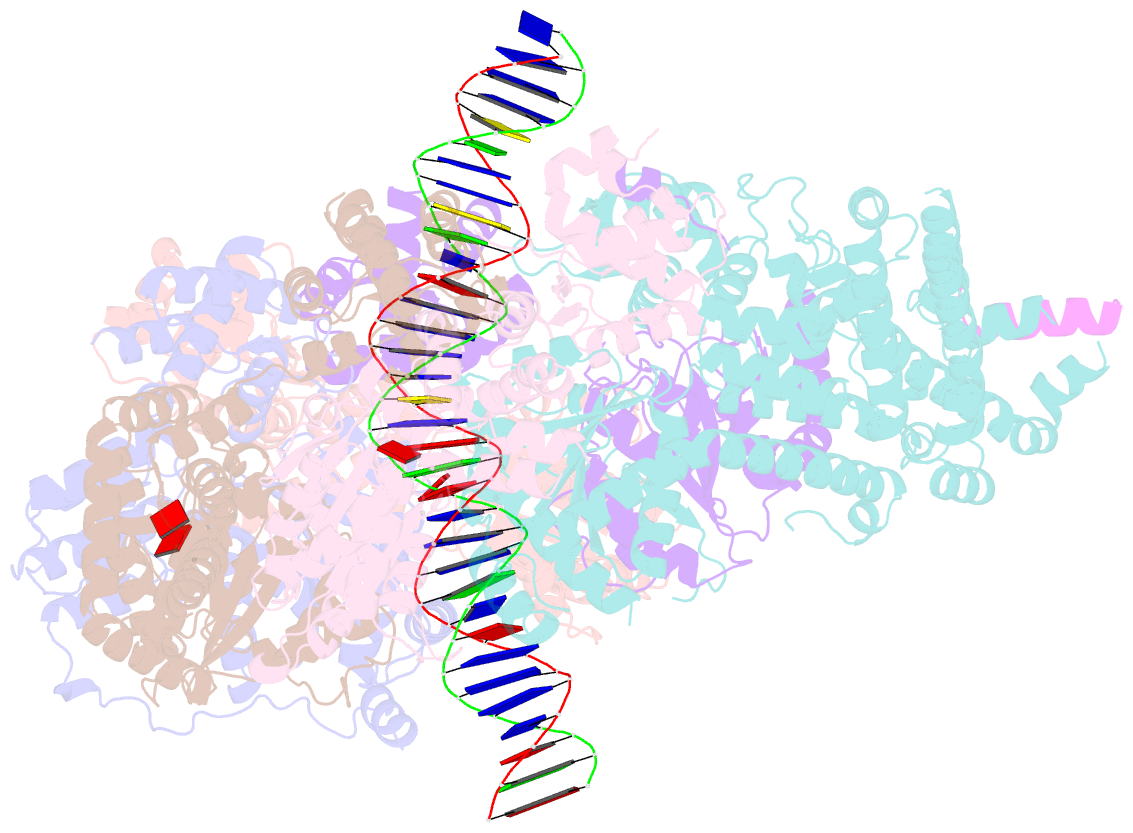
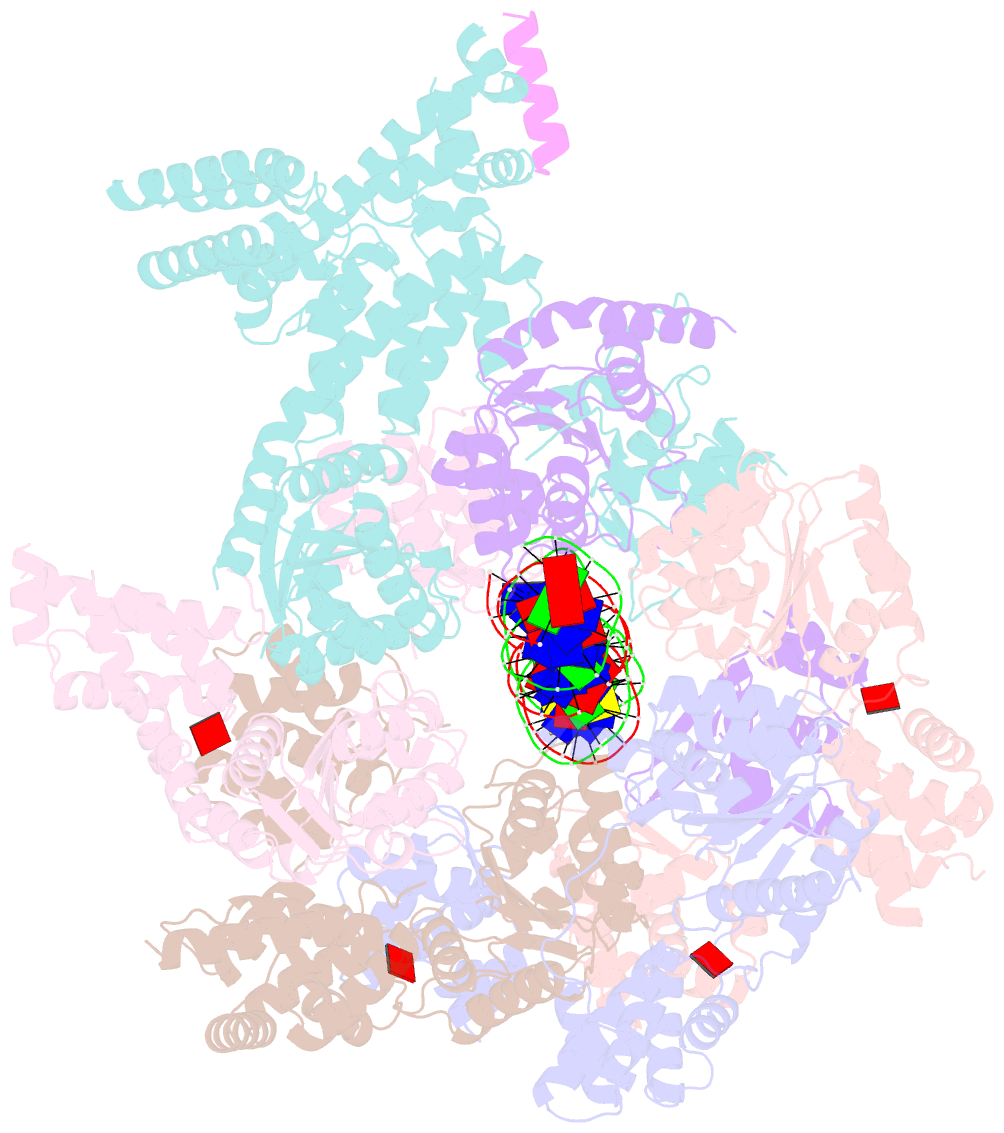
Summary information and primary citation
- PDB-id
- 7jgr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- cryo-EM (3.9 Å)
- Summary
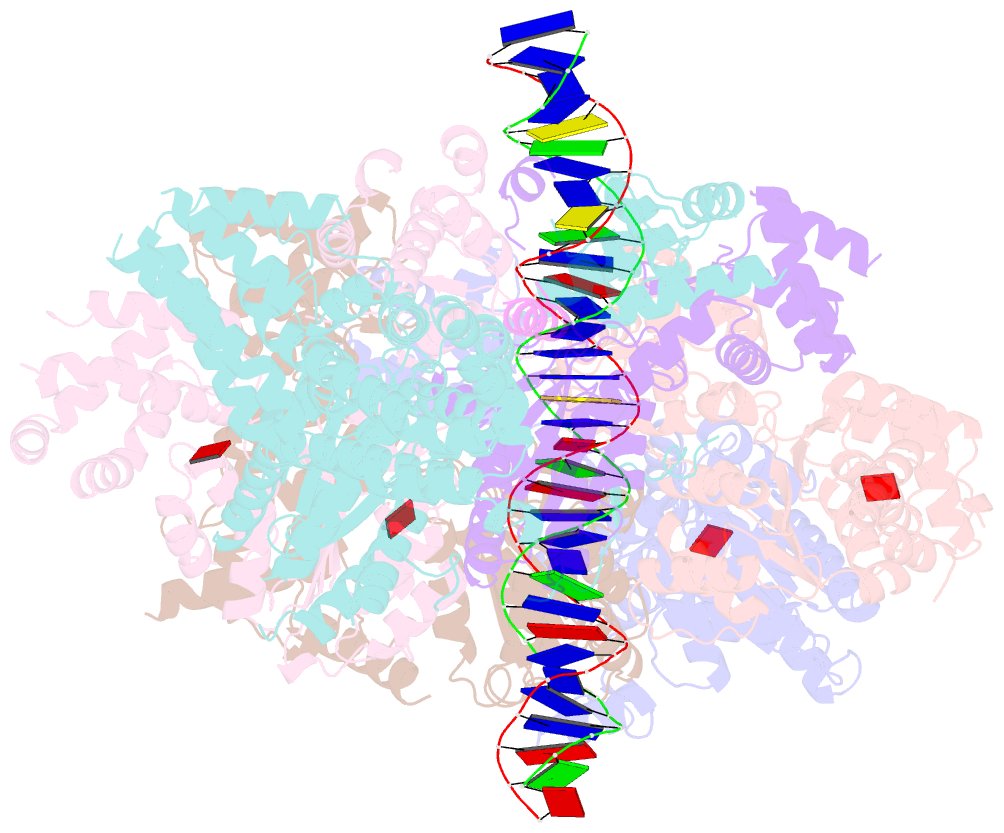
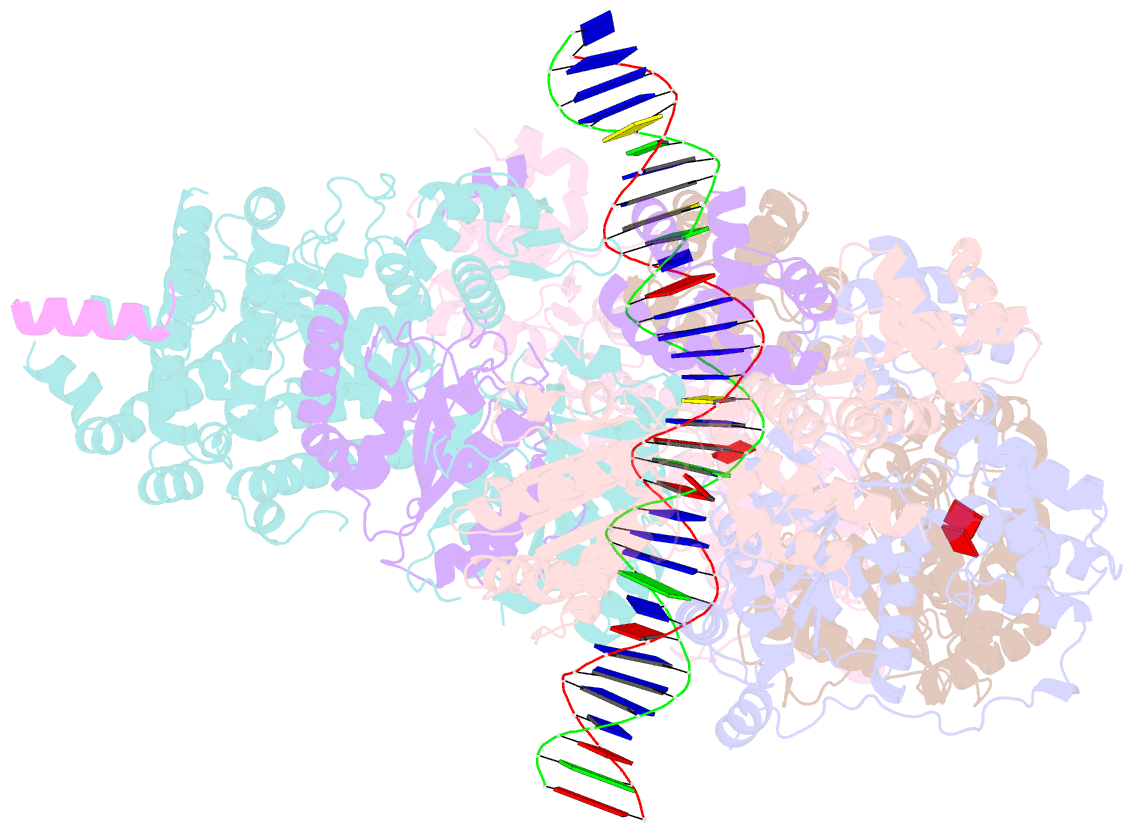
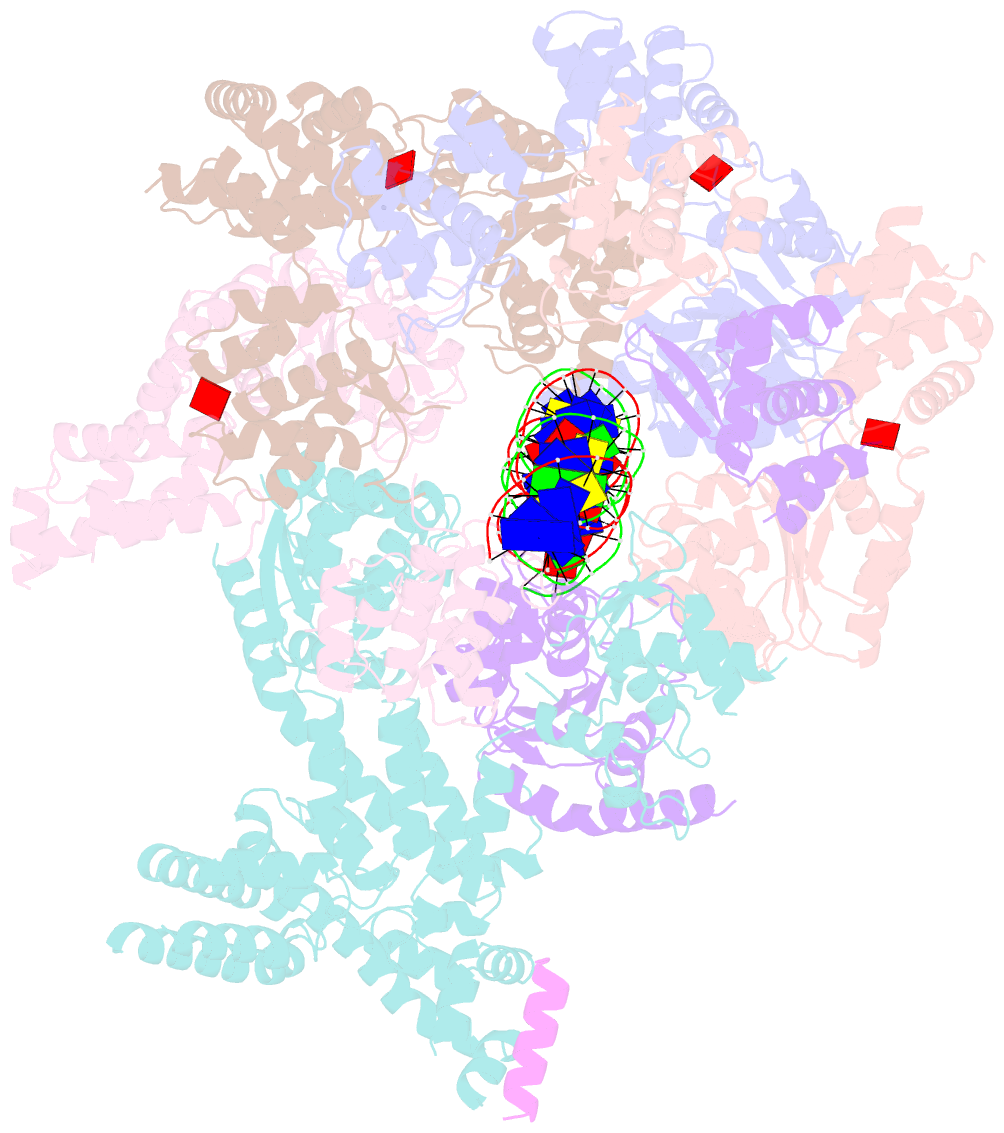
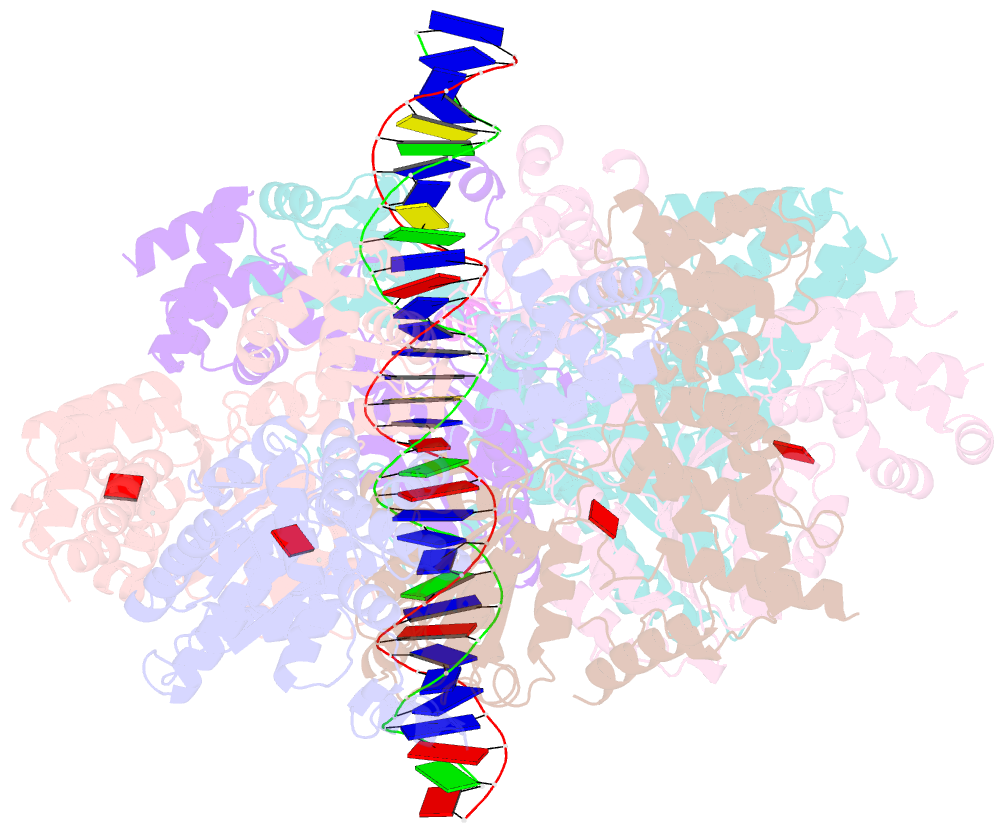
- Structure of drosophila orc bound to DNA (84 bp) and cdc6
- Reference
- Schmidt JM, Bleichert F (2020): "Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6." Nat Commun, 11, 4263. doi: 10.1038/s41467-020-18067-7.
- Abstract
- Eukaryotic DNA replication initiation relies on the origin recognition complex (ORC), a DNA-binding ATPase that loads the Mcm2-7 replicative helicase onto replication origins. Here, we report cryo-electron microscopy (cryo-EM) structures of DNA-bound Drosophila ORC with and without the co-loader Cdc6. These structures reveal that Orc1 and Orc4 constitute the primary DNA binding site in the ORC ring and cooperate with the winged-helix domains to stabilize DNA bending. A loop region near the catalytic Walker B motif of Orc1 directly contacts DNA, allosterically coupling DNA binding to ORC's ATPase site. Correlating structural and biochemical data show that DNA sequence modulates DNA binding and remodeling by ORC, and that DNA bending promotes Mcm2-7 loading in vitro. Together, these findings explain the distinct DNA sequence-dependencies of metazoan and S. cerevisiae initiators in origin recognition and support a model in which DNA geometry and bendability contribute to Mcm2-7 loading site selection in metazoans.