Summary information and primary citation
- PDB-id
- 7jk1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.62 Å)
- Summary
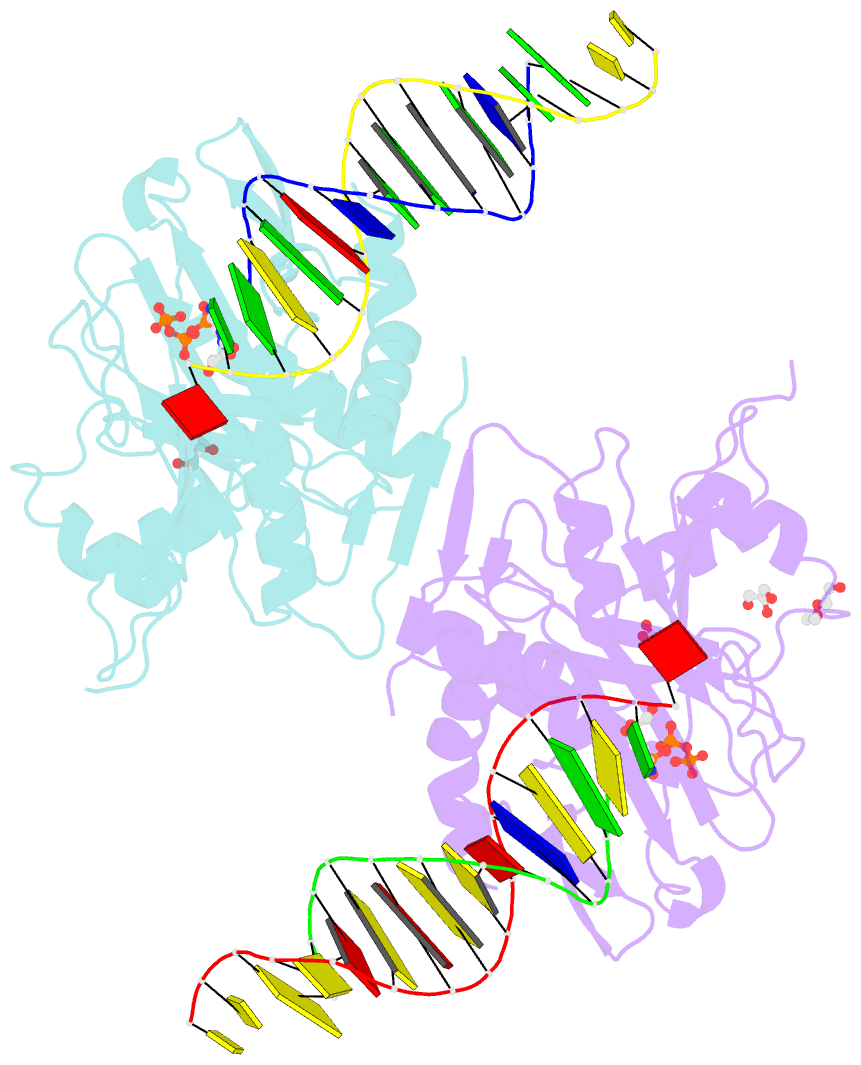
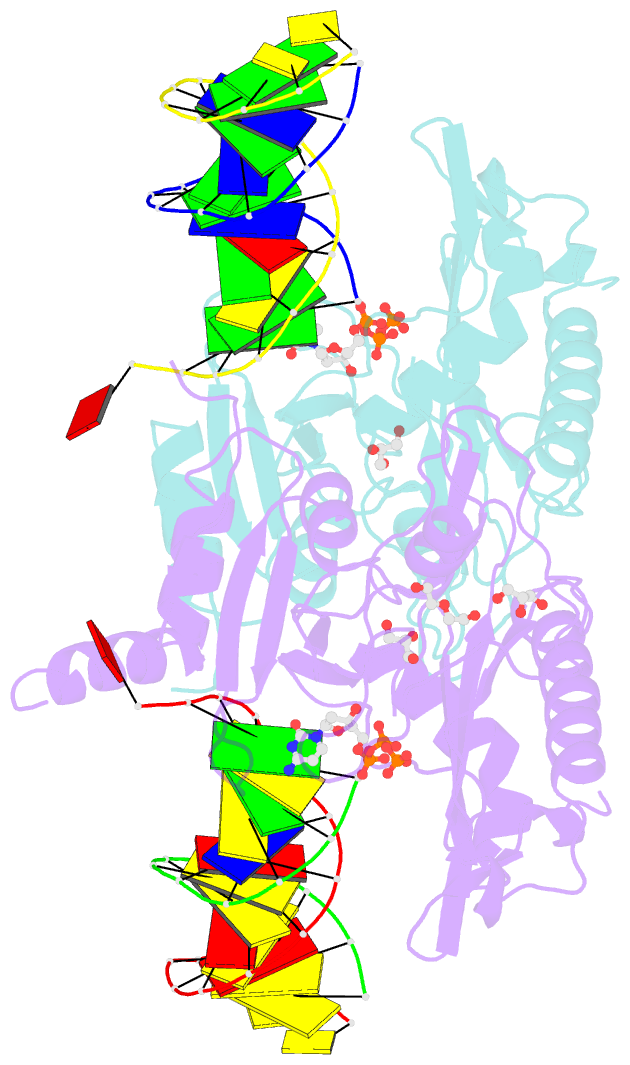
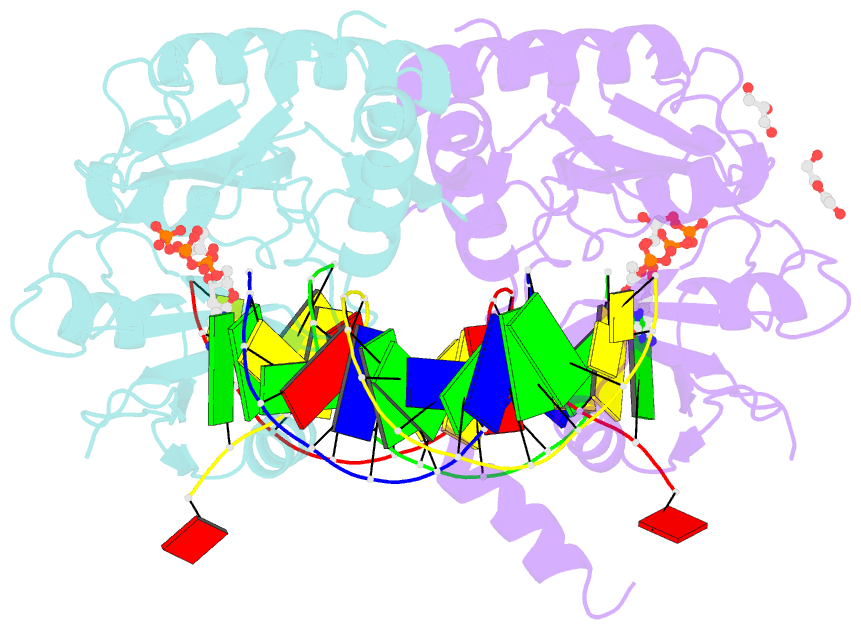
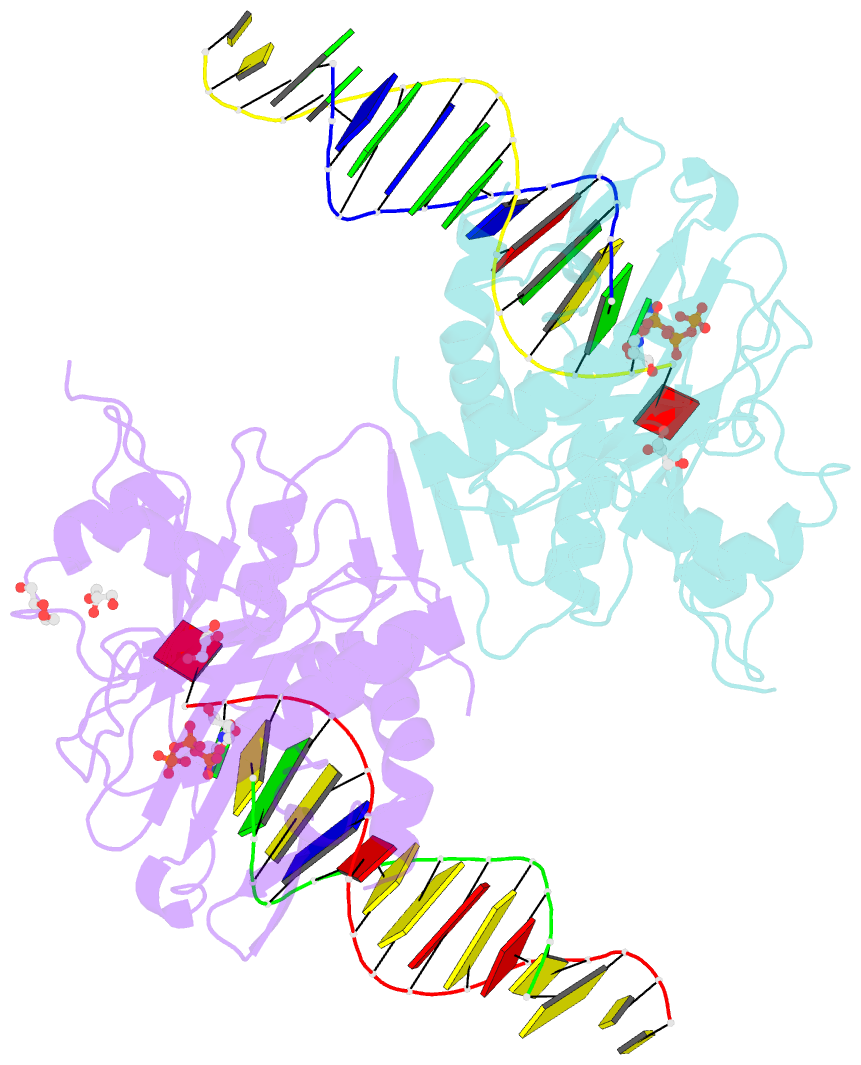
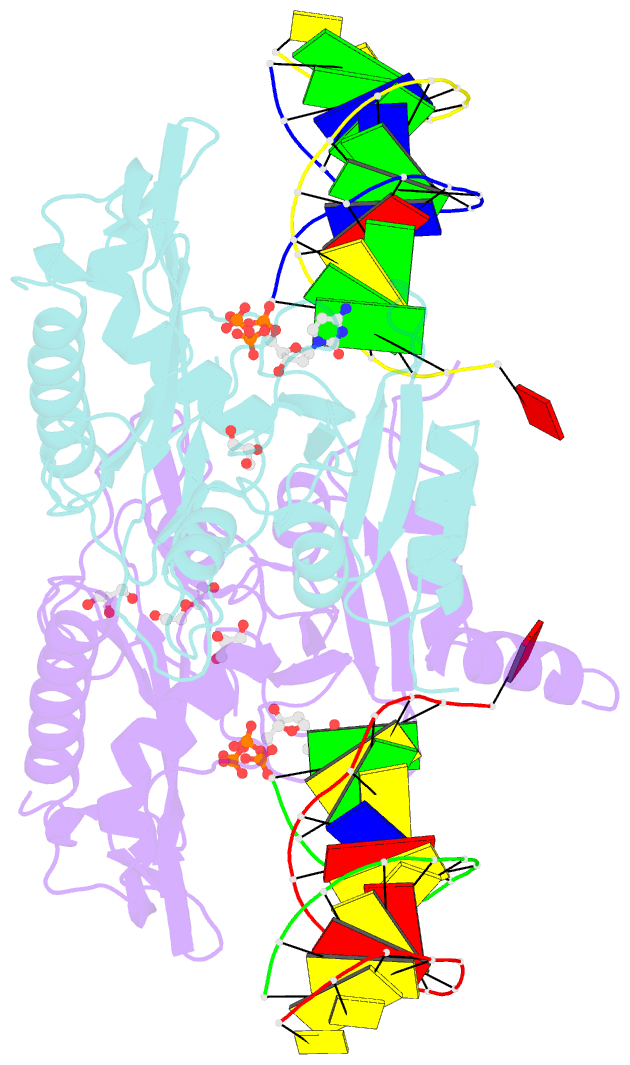
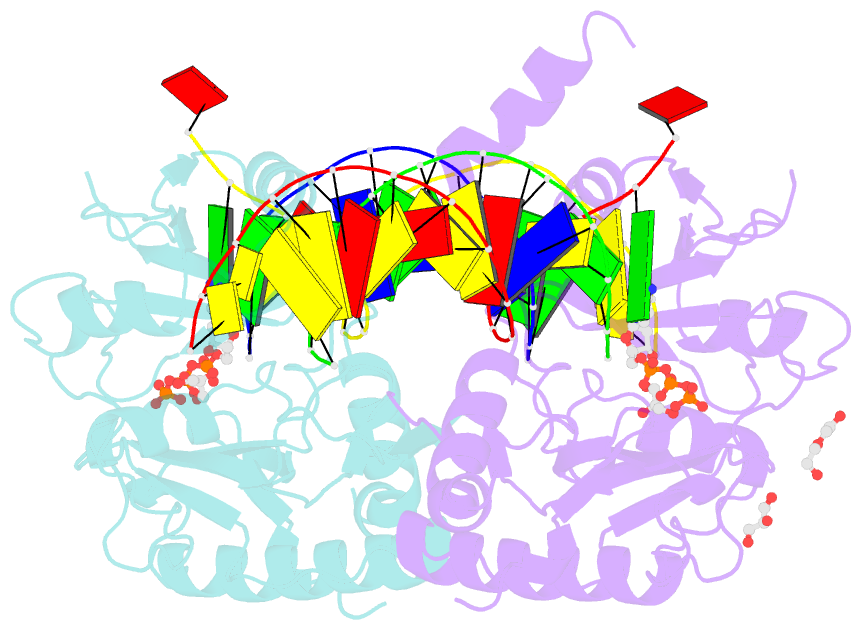
- Human primpol inserting correct dctp opposite the 8-oxoguanine lesion
- Reference
- Rechkoblit O, Johnson RE, Gupta YK, Prakash L, Prakash S, Aggarwal AK (2021): "Structural basis of DNA synthesis opposite 8-oxoguanine by human PrimPol primase-polymerase." Nat Commun, 12, 4020. doi: 10.1038/s41467-021-24317-z.
- Abstract
- PrimPol is a human DNA polymerase-primase that localizes to mitochondria and nucleus and bypasses the major oxidative lesion 7,8-dihydro-8-oxoguanine (oxoG) via translesion synthesis, in mostly error-free manner. We present structures of PrimPol insertion complexes with a DNA template-primer and correct dCTP or erroneous dATP opposite the lesion, as well as extension complexes with C or A as a 3'-terminal primer base. We show that during the insertion of C and extension from it, the active site is unperturbed, reflecting the readiness of PrimPol to accommodate oxoG(anti). The misinsertion of A opposite oxoG(syn) also does not alter the active site, and is likely less favorable due to lower thermodynamic stability of the oxoG(syn)•A base-pair. During the extension step, oxoG(syn) induces an opening of its base-pair with A or misalignment of the 3'-A primer terminus. Together, the structures show how PrimPol accurately synthesizes DNA opposite oxidatively damaged DNA in human cells.