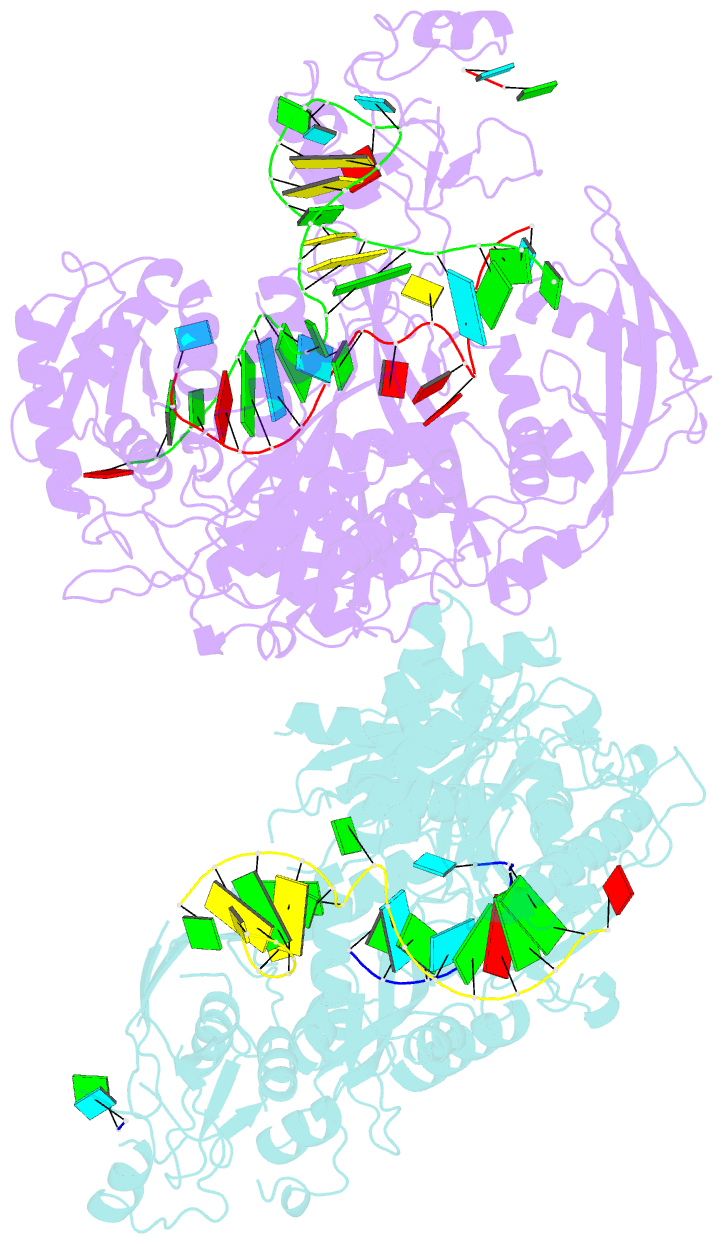
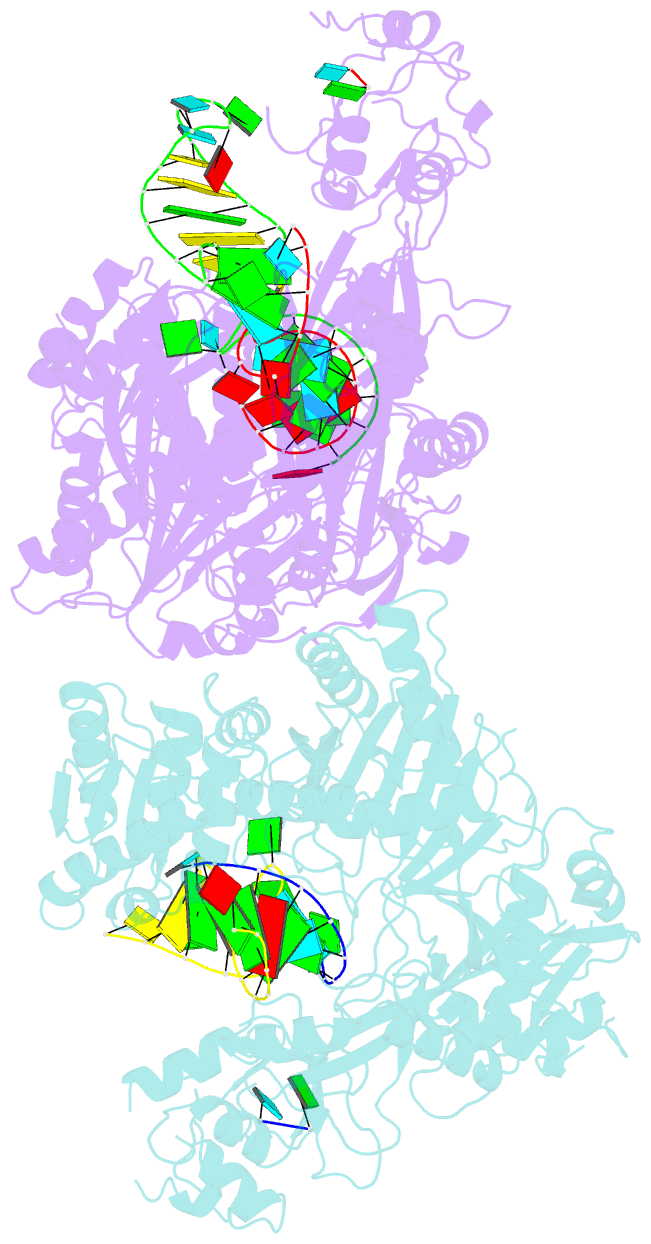
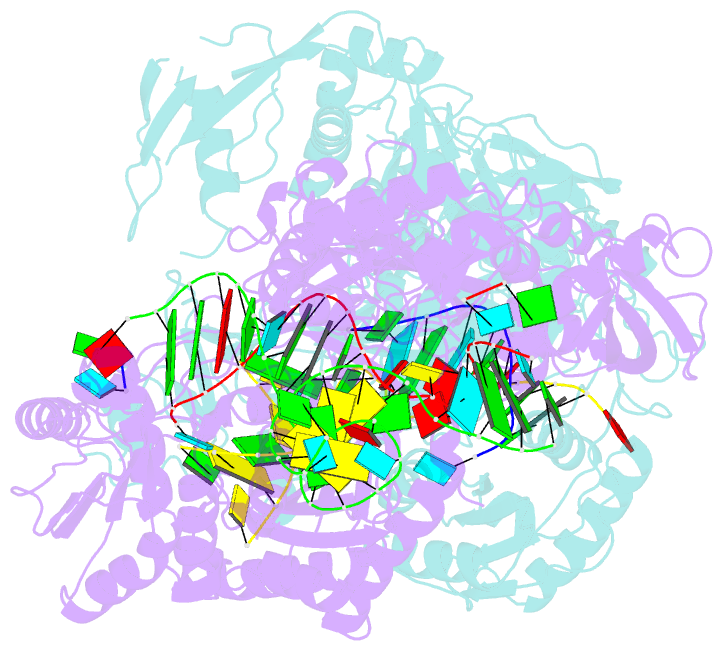
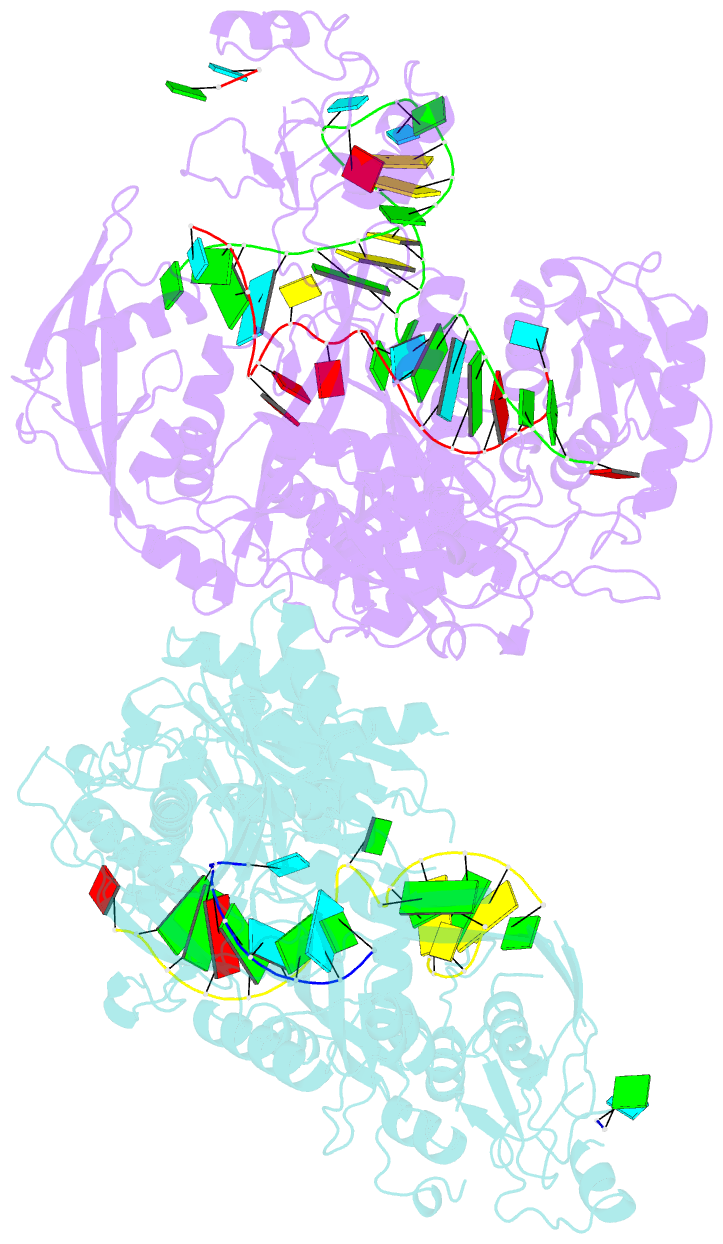
Summary information and primary citation
- PDB-id
- 7ki3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.0 Å)
- Summary
- Human argonaute2:mir-122 bound to the hcv genotype 1a site-1 RNA
- Reference
- Gebert LFR, Law M, MacRae IJ (2021): "A structured RNA motif locks Argonaute2:miR-122 onto the 5' end of the HCV genome." Nat Commun, 12, 6836. doi: 10.1038/s41467-021-27177-9.
- Abstract
- microRNAs (miRNAs) form regulatory networks in metazoans. Viruses engage miRNA networks in numerous ways, with Flaviviridae members exploiting direct interactions of their RNA genomes with host miRNAs. For hepatitis C virus (HCV), binding of liver-abundant miR-122 stabilizes the viral RNA and regulates viral translation. Here, we investigate the structural basis for these activities, taking into consideration that miRNAs function in complex with Argonaute (Ago) proteins. The crystal structure of the Ago2:miR-122:HCV complex reveals a structured RNA motif that traps Ago2 on the viral RNA, masking its 5' end from enzymatic attack. The trapped Ago2 can recruit host factor PCBP2, implicated in viral translation, while binding of a second Ago2:miR-122 competes with PCBP2, creating a potential molecular switch for translational control. Combined results reveal a viral RNA structure that modulates Ago2:miR-122 dynamics and repurposes host proteins to generate a functional analog of the mRNA cap-binding complex.