Summary information and primary citation
- PDB-id
- 7kii; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (1.3 Å)
- Summary
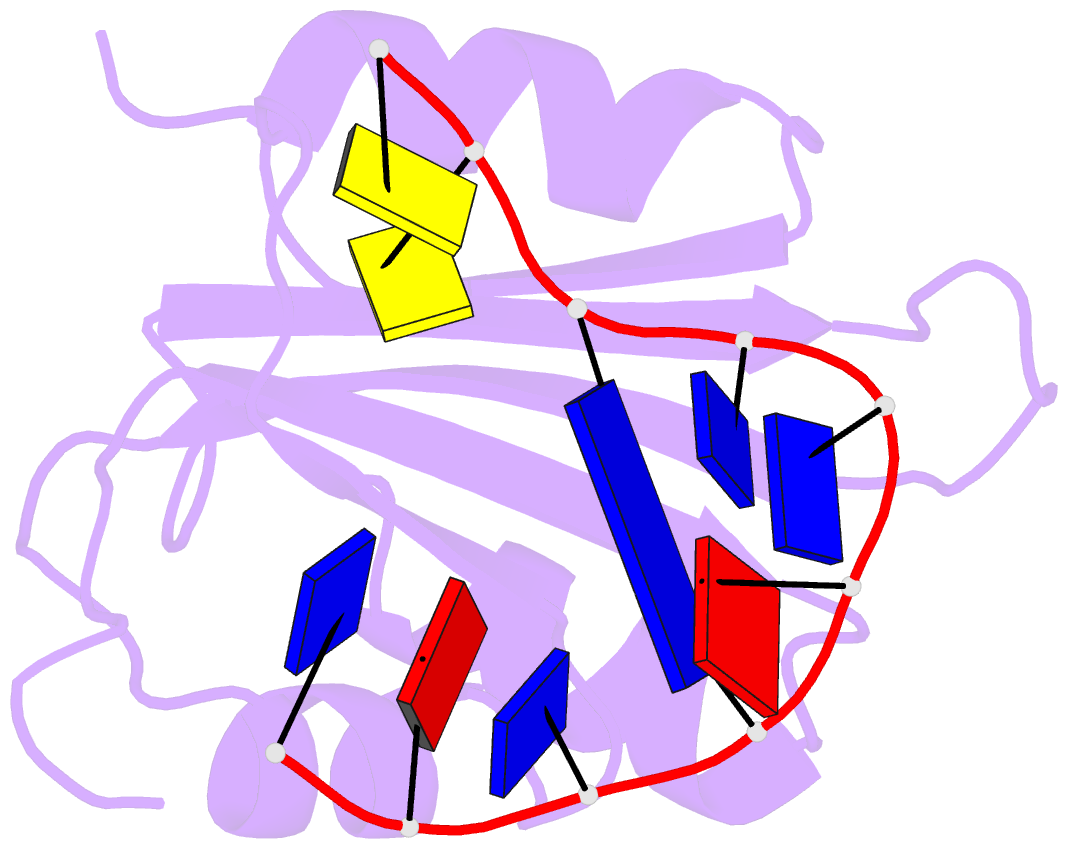
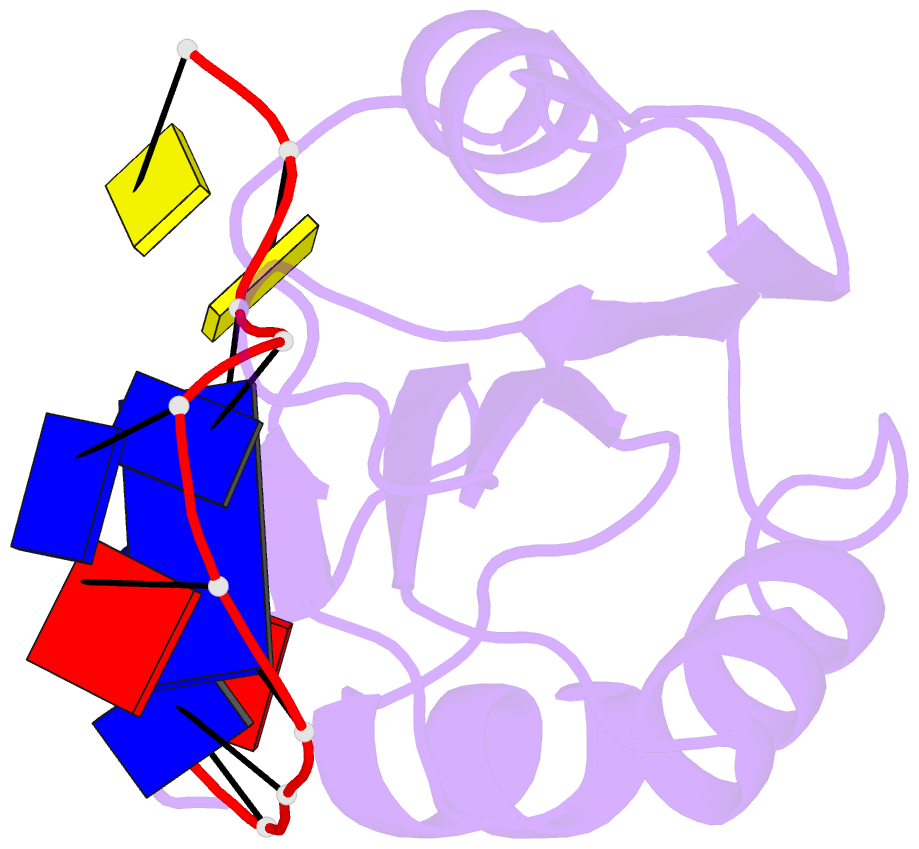
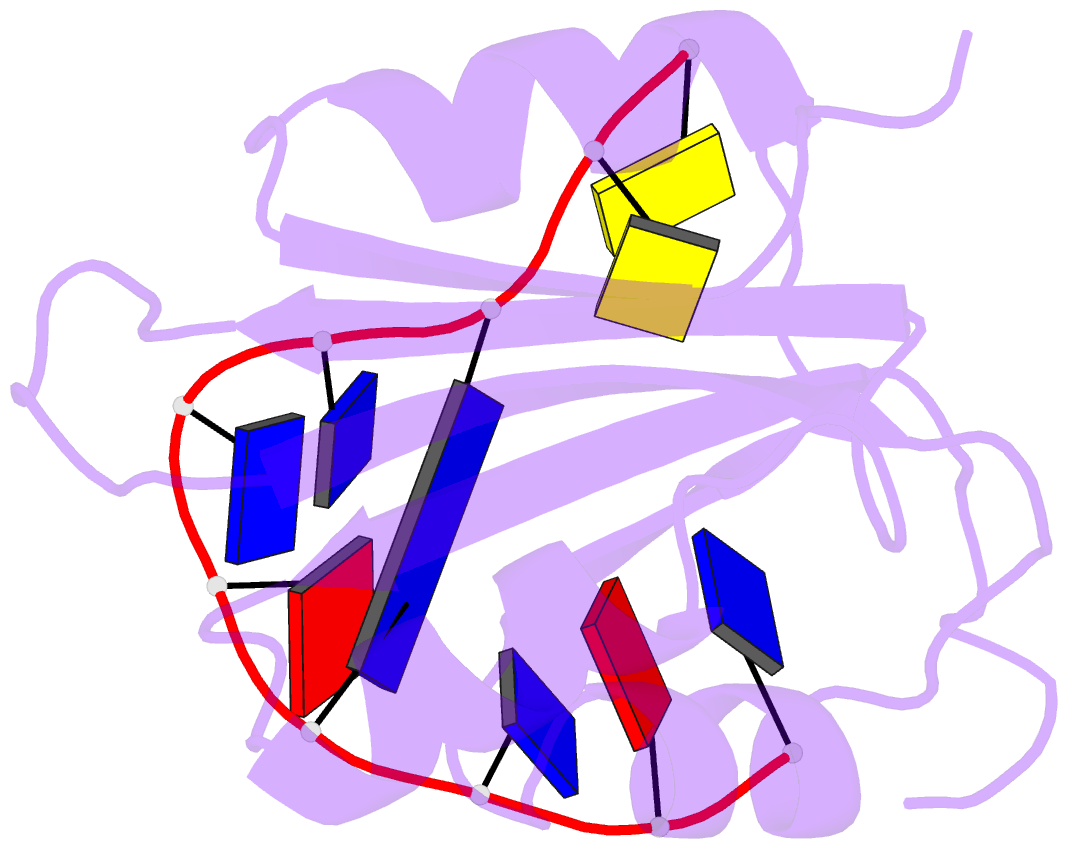
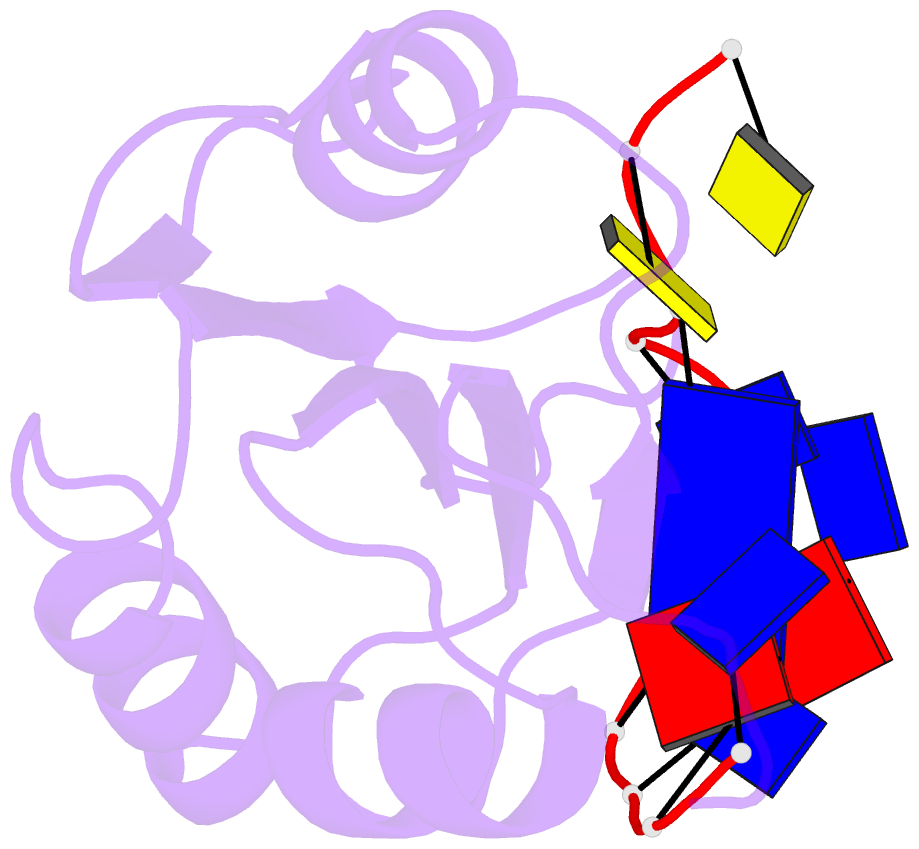
- Muscovy duck circovirus rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site and mn2+
- Reference
- Smiley AT, Tompkins KJ, Pawlak MR, Krueger AJ, Evans 3rd RL, Shi K, Aihara H, Gordon WR (2023): "Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases." Mbio, 14, e0258722. doi: 10.1128/mbio.02587-22.
- Abstract
- Replication-initiating HUH endonucleases (Reps) are sequence-specific nucleases that cleave and rejoin single-stranded DNA (ssDNA) during rolling-circle replication. These functions are mediated by covalent linkage of the Rep to its substrate post cleavage. Here, we describe the structures of the endonuclease domain from the Muscovy duck circovirus Rep in complex with its cognate ssDNA 10-mer with and without manganese in the active site. Structural and functional analyses demonstrate that divalent cations play both catalytic and structural roles in Reps by polarizing and positioning their substrate. Further structural comparisons highlight the importance of an intramolecular substrate Watson-Crick (WC) base pairing between the -4 and +1 positions. Subsequent kinetic and functional analyses demonstrate a functional dependency on WC base pairing between these positions regardless of the pair's identity (i.e., A·T, T·A, G·C, or C·G), highlighting a structural specificity for substrate interaction. Finally, considering how well WC swaps were tolerated in vitro, we sought to determine to what extent the canonical -4T·+1A pairing is conserved in circular Rep-encoding single-stranded DNA viruses and found evidence of noncanonical pairings in a minority of these genomes. Altogether, our data suggest that substrate intramolecular WC base pairing is a universal requirement for separation and reunion of ssDNA in Reps.