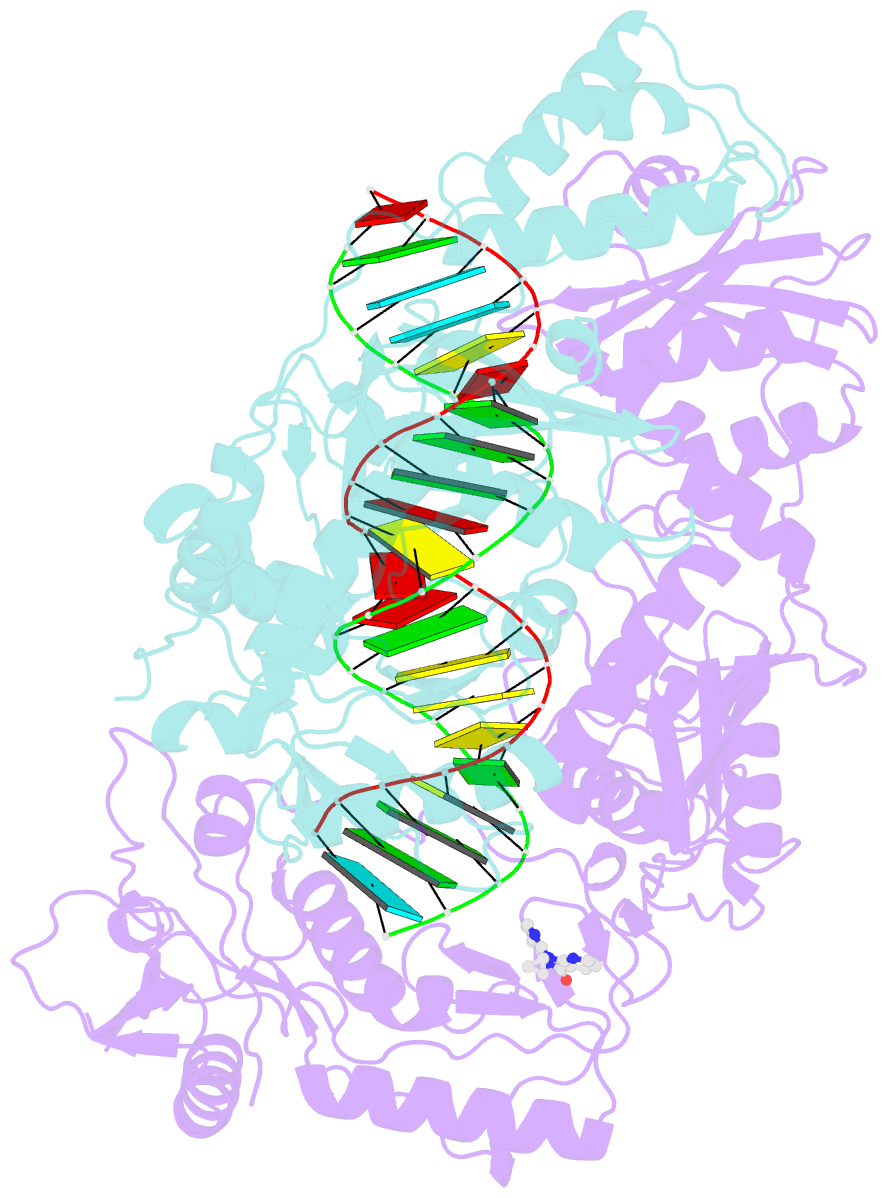
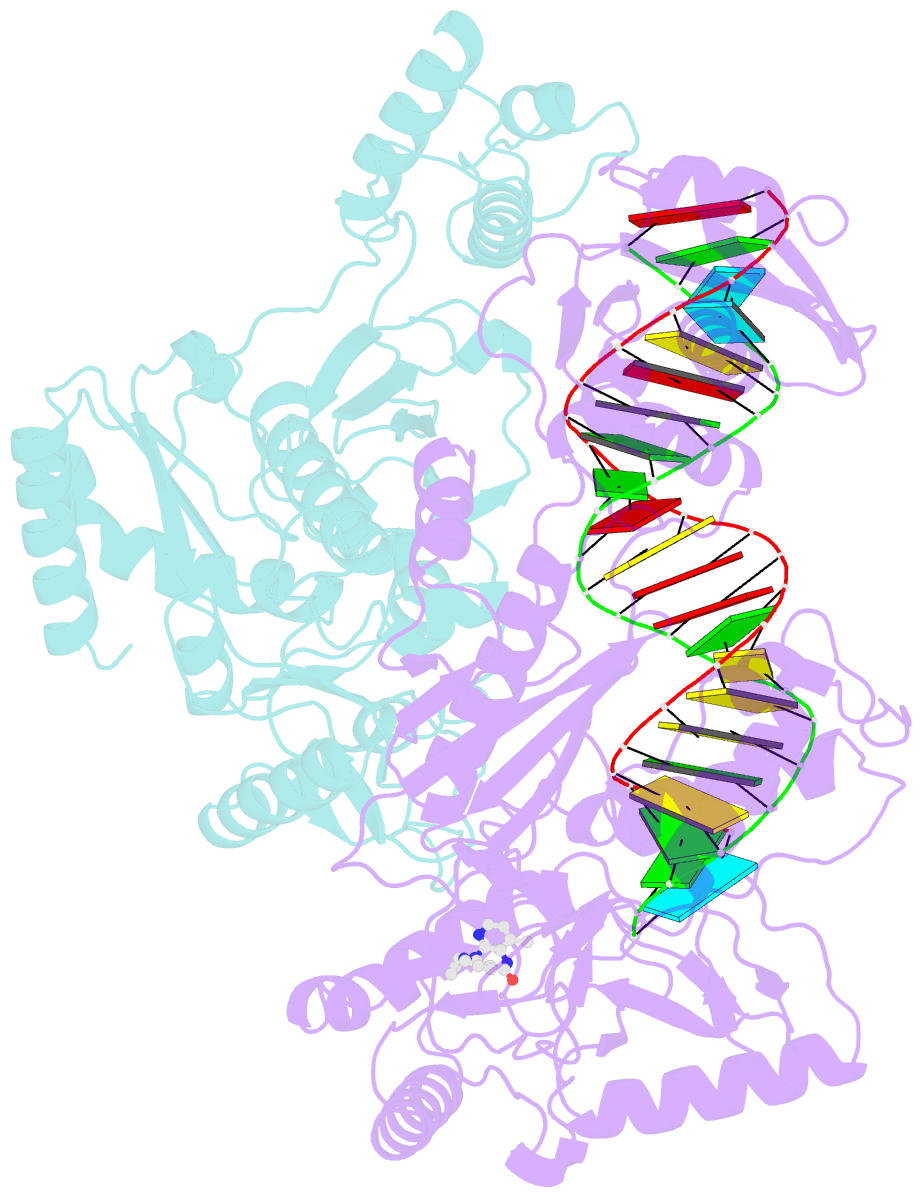
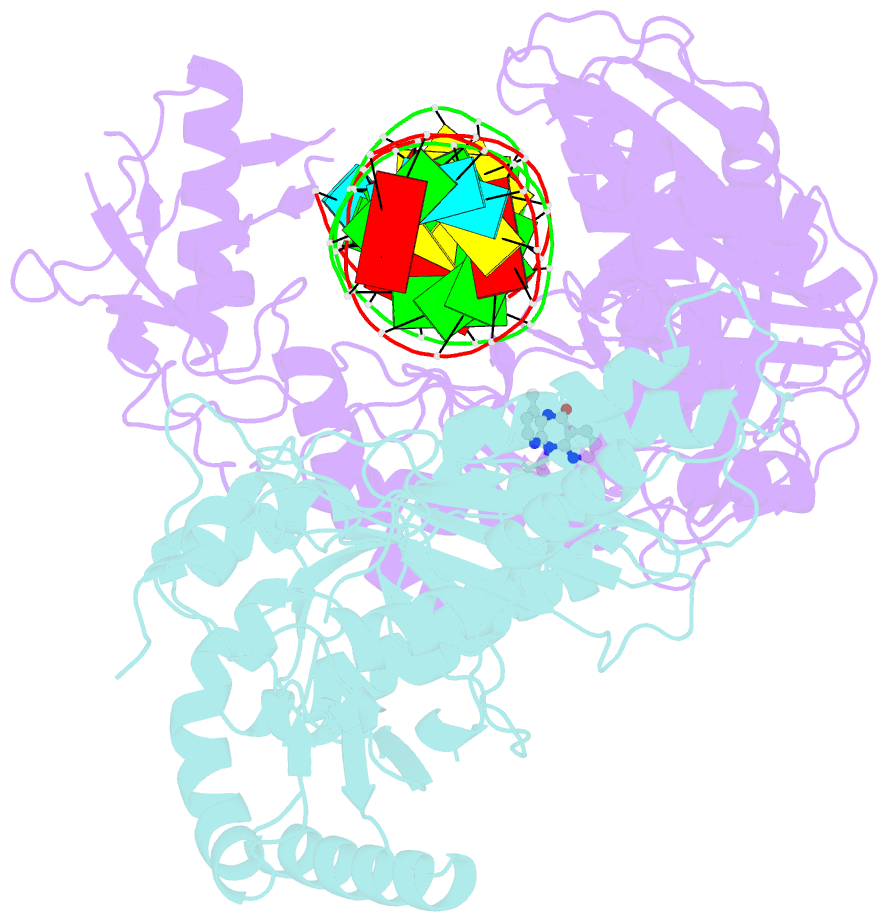
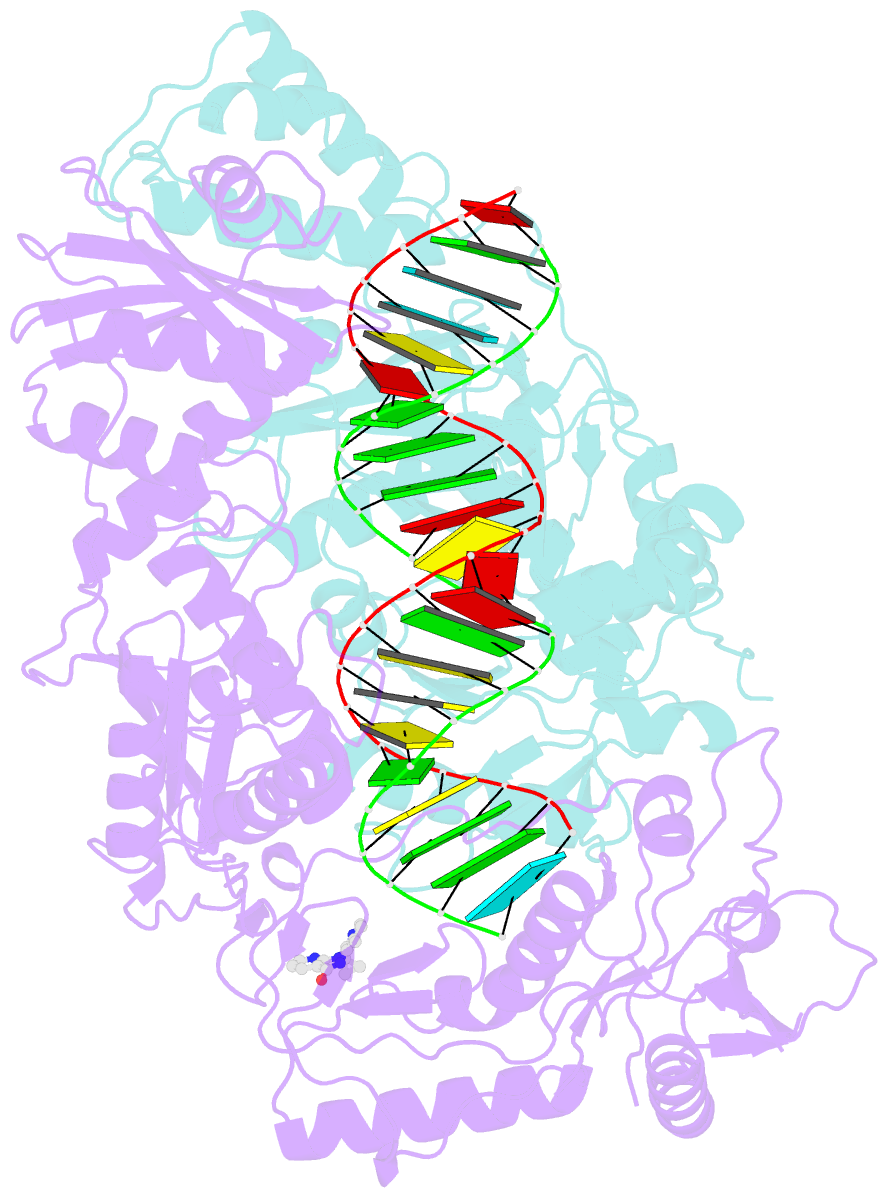
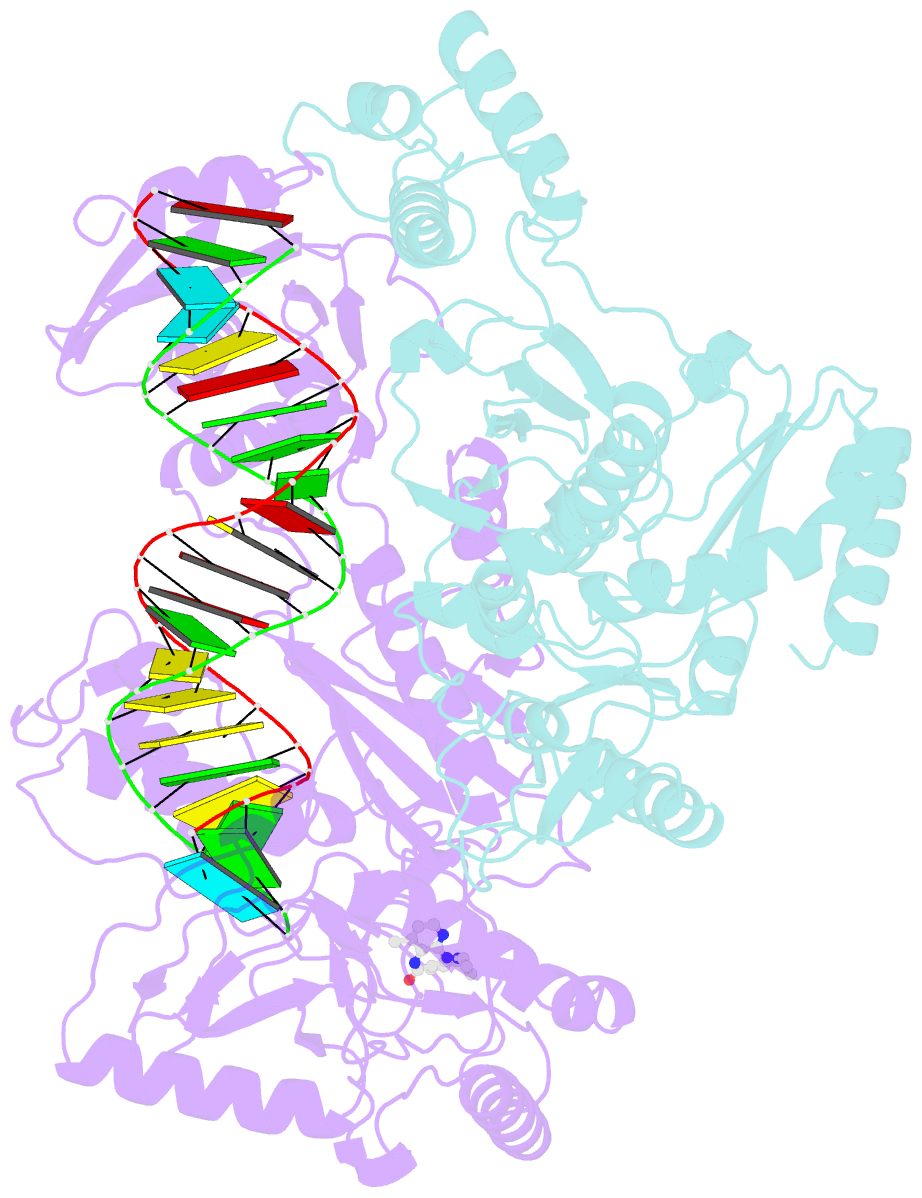
Summary information and primary citation
- PDB-id
- 7kjx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- cryo-EM (3.1 Å)
- Summary
- Structure of hiv-1 reverse transcriptase initiation complex core with nevirapine
- Reference
- Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV (2021): "High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs." Nat Commun, 12, 2500. doi: 10.1038/s41467-021-22628-9.
- Abstract
- Reverse transcription of the HIV-1 viral RNA genome (vRNA) is an integral step in virus replication. Upon viral entry, HIV-1 reverse transcriptase (RT) initiates from a host tRNALys3 primer bound to the vRNA genome and is the target of key antivirals, such as non-nucleoside reverse transcriptase inhibitors (NNRTIs). Initiation proceeds slowly with discrete pausing events along the vRNA template. Despite prior medium-resolution structural characterization of reverse transcriptase initiation complexes (RTICs), higher-resolution structures of the RTIC are needed to understand the molecular mechanisms that underlie initiation. Here we report cryo-EM structures of the core RTIC, RTIC-nevirapine, and RTIC-efavirenz complexes at 2.8, 3.1, and 2.9 Å, respectively. In combination with biochemical studies, these data suggest a basis for rapid dissociation kinetics of RT from the vRNA-tRNALys3 initiation complex and reveal a specific structural mechanism of nucleic acid conformational stabilization during initiation. Finally, our results show that NNRTIs inhibit the RTIC and exacerbate discrete pausing during early reverse transcription.