Summary information and primary citation
- PDB-id
- 7kro; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-hydrolase-RNA
- Method
- cryo-EM (3.6 Å)
- Summary
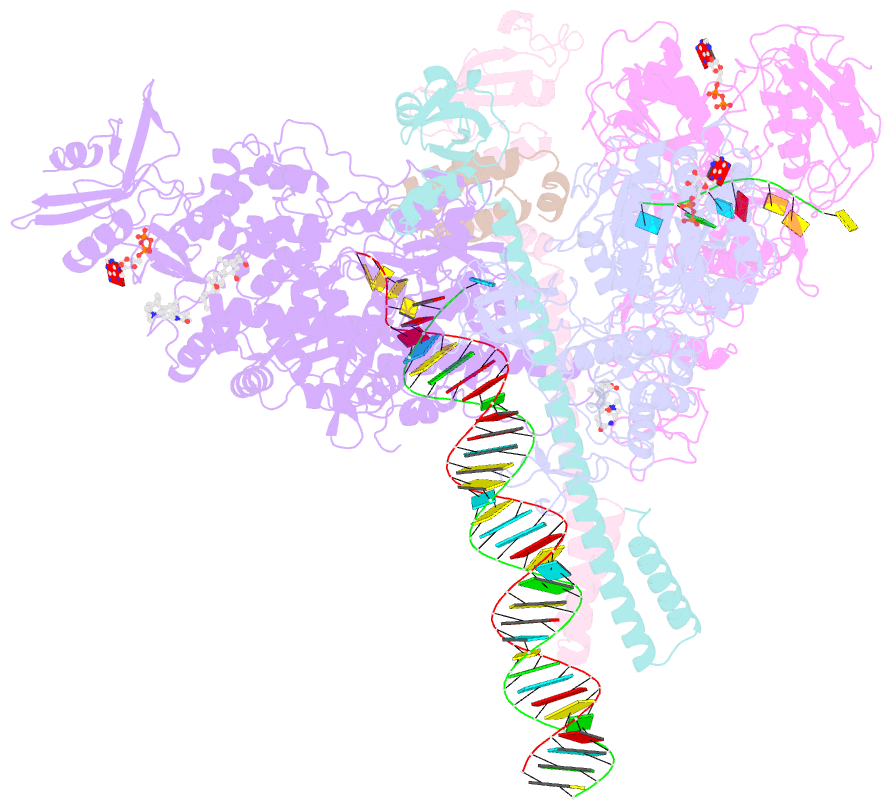
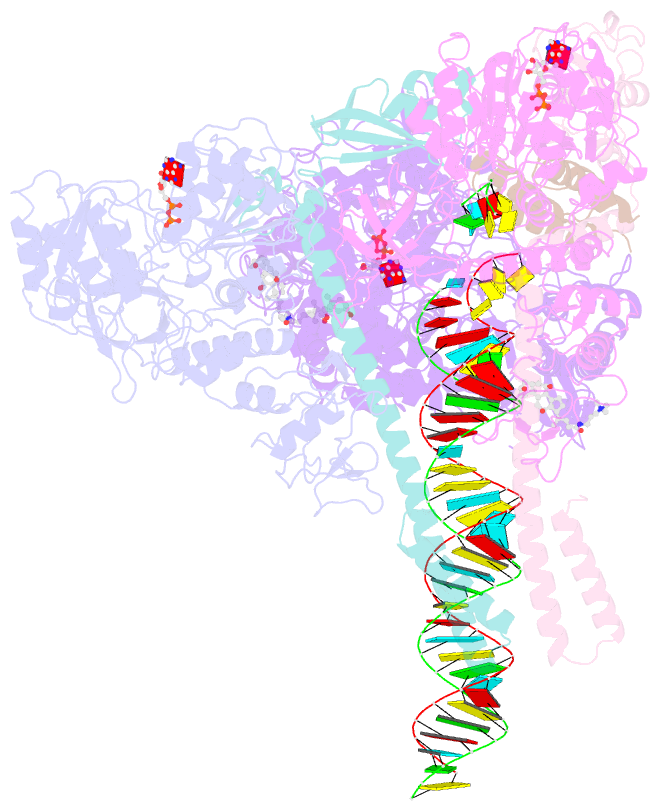
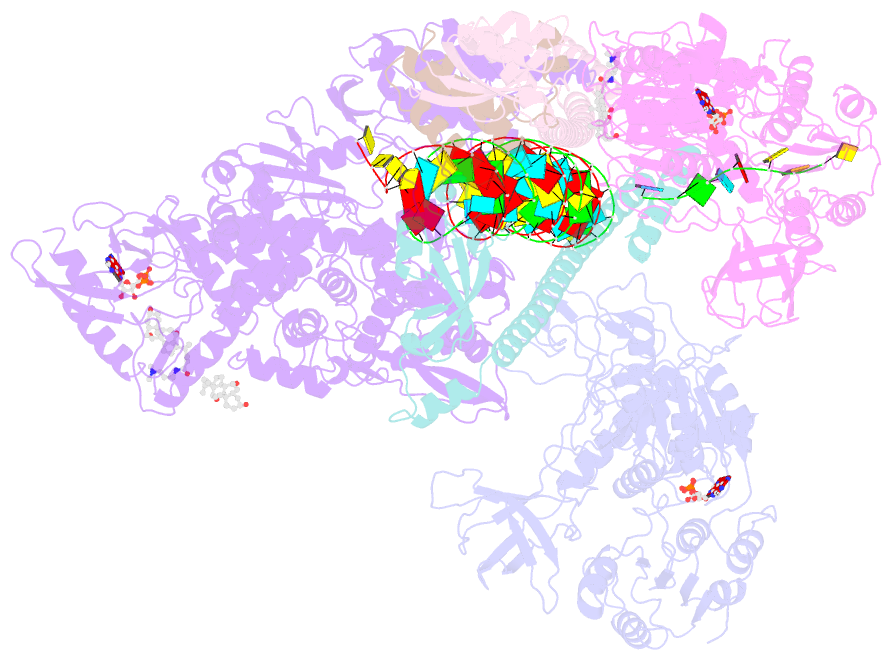
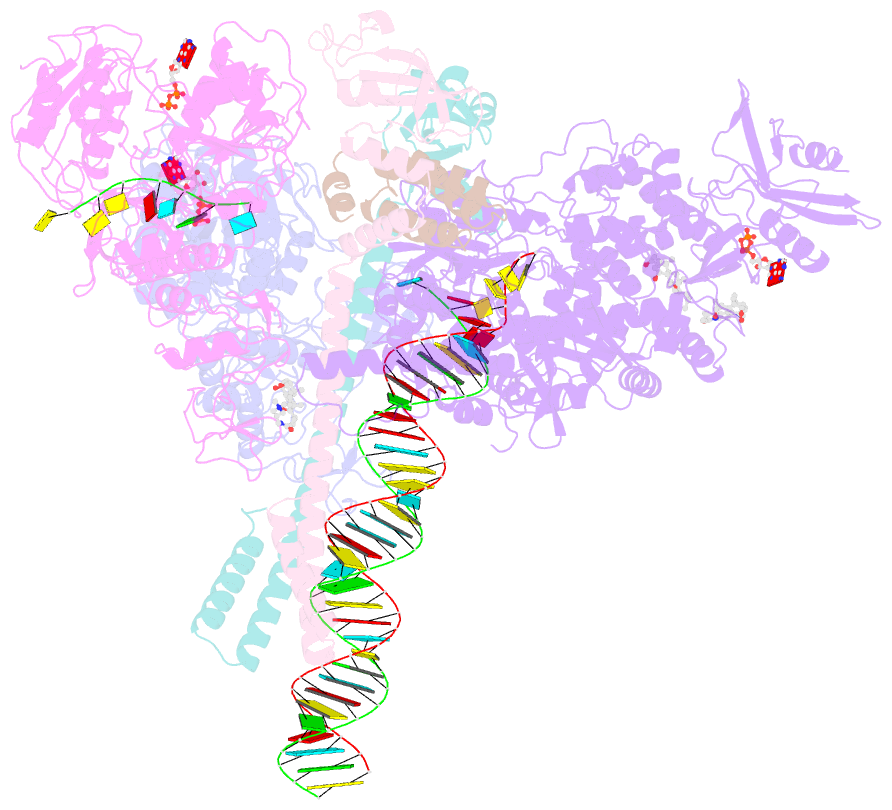
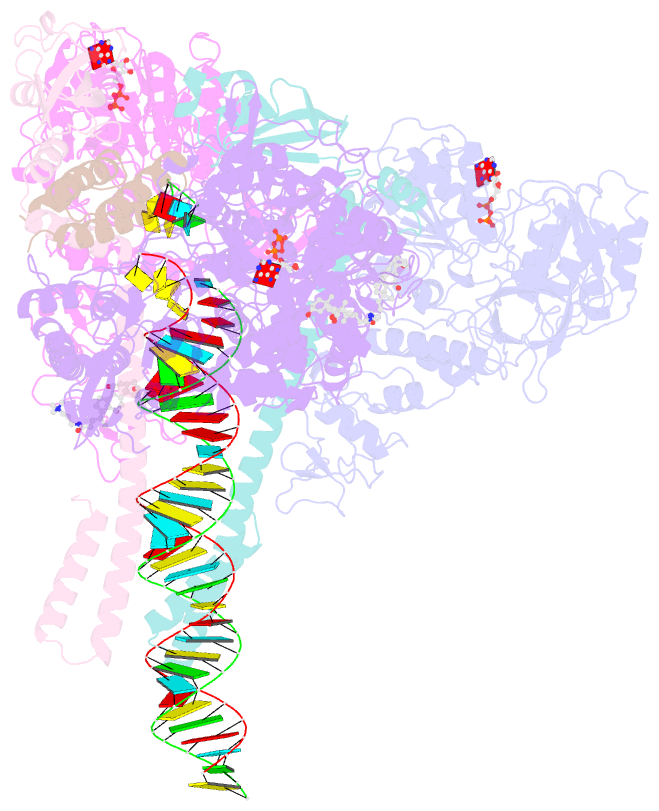
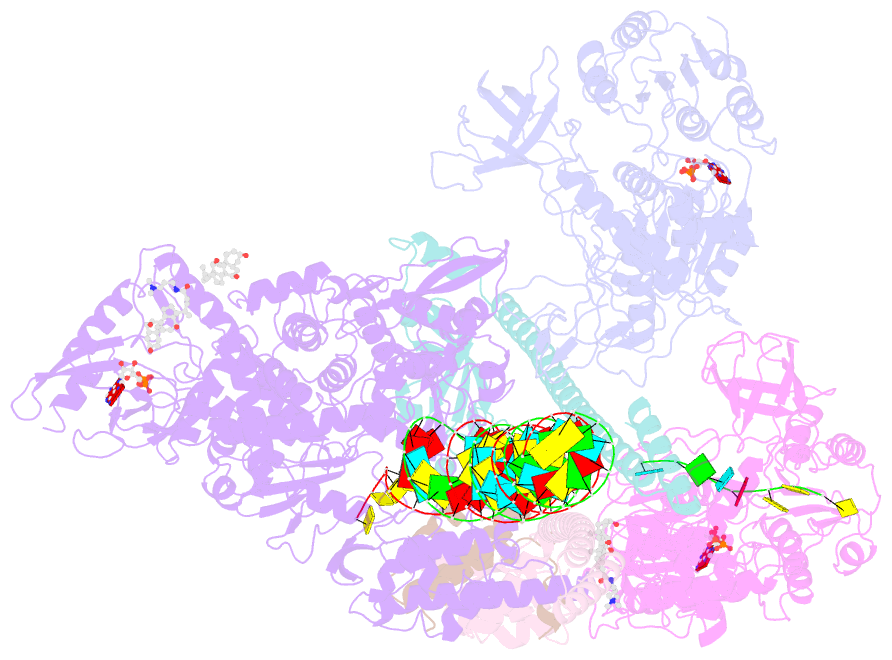
- Structure of sars-cov-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-btc
- Reference
- Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA (2021): "Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex." Proc.Natl.Acad.Sci.USA, 118. doi: 10.1073/pnas.2102516118.
- Abstract
- Backtracking, the reverse motion of the transcriptase enzyme on the nucleic acid template, is a universal regulatory feature of transcription in cellular organisms but its role in viruses is not established. Here we present evidence that backtracking extends into the viral realm, where backtracking by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) may aid viral transcription and replication. Structures of SARS-CoV-2 RdRp bound to the essential nsp13 helicase and RNA suggested the helicase facilitates backtracking. We use cryo-electron microscopy, RNA-protein cross-linking, and unbiased molecular dynamics simulations to characterize SARS-CoV-2 RdRp backtracking. The results establish that the single-stranded 3' segment of the product RNA generated by backtracking extrudes through the RdRp nucleoside triphosphate (NTP) entry tunnel, that a mismatched nucleotide at the product RNA 3' end frays and enters the NTP entry tunnel to initiate backtracking, and that nsp13 stimulates RdRp backtracking. Backtracking may aid proofreading, a crucial process for SARS-CoV-2 resistance against antivirals.