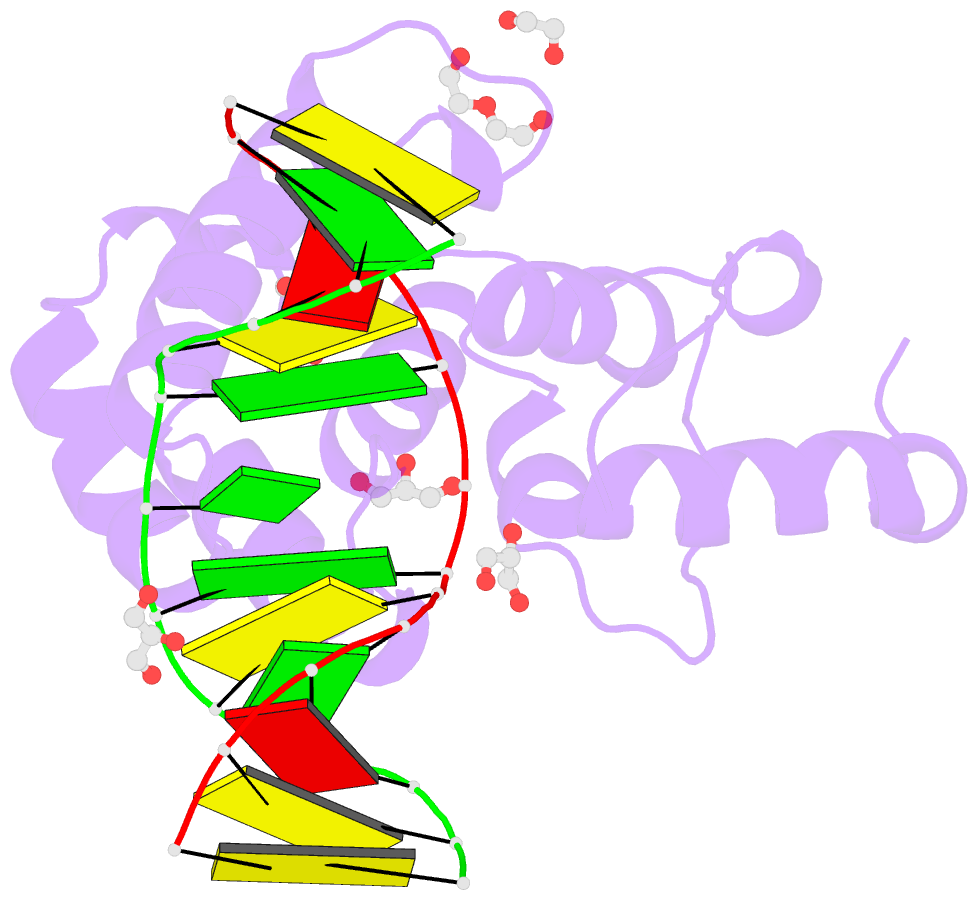
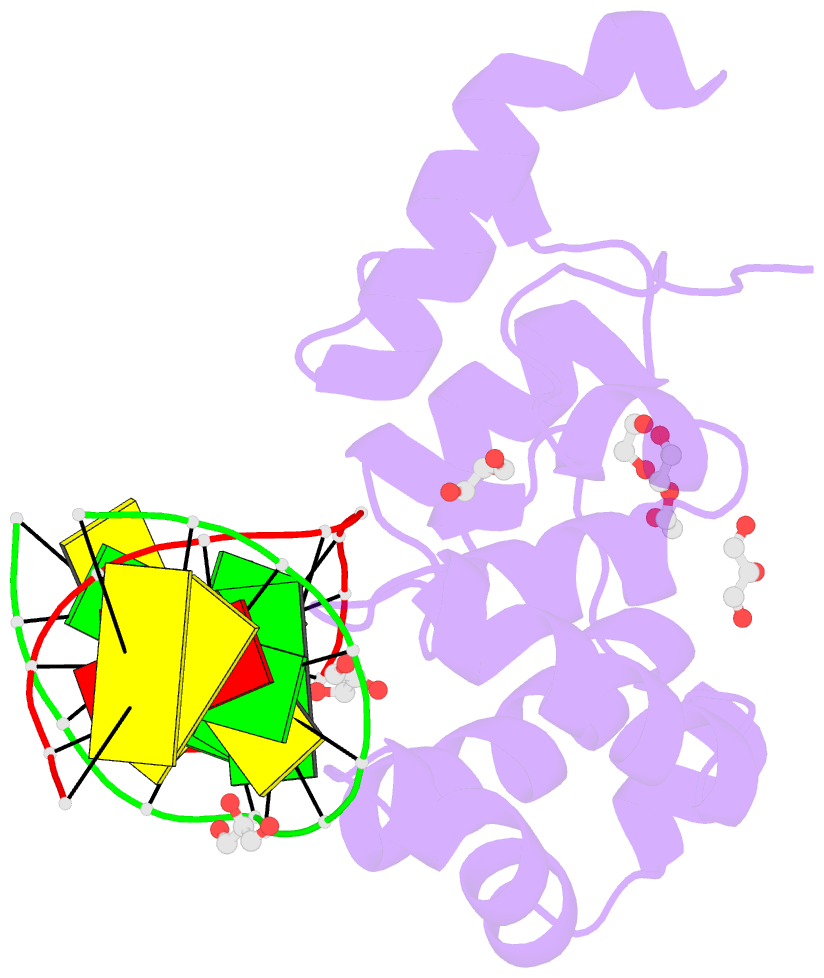
Summary information and primary citation
- PDB-id
- 7kzg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.68 Å)
- Summary
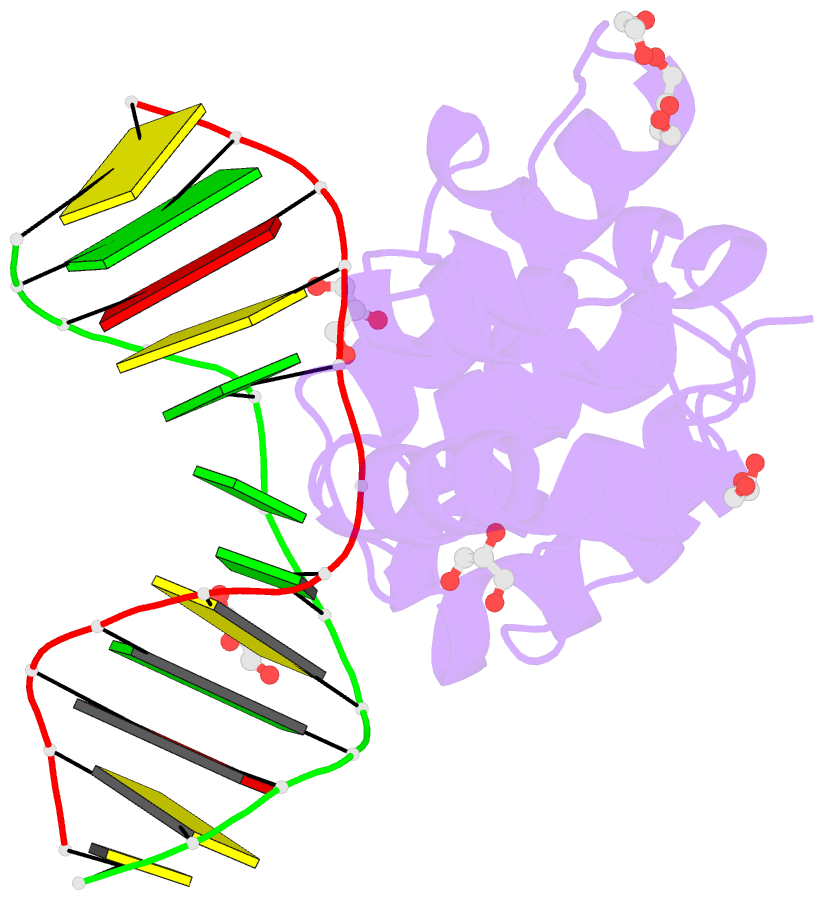
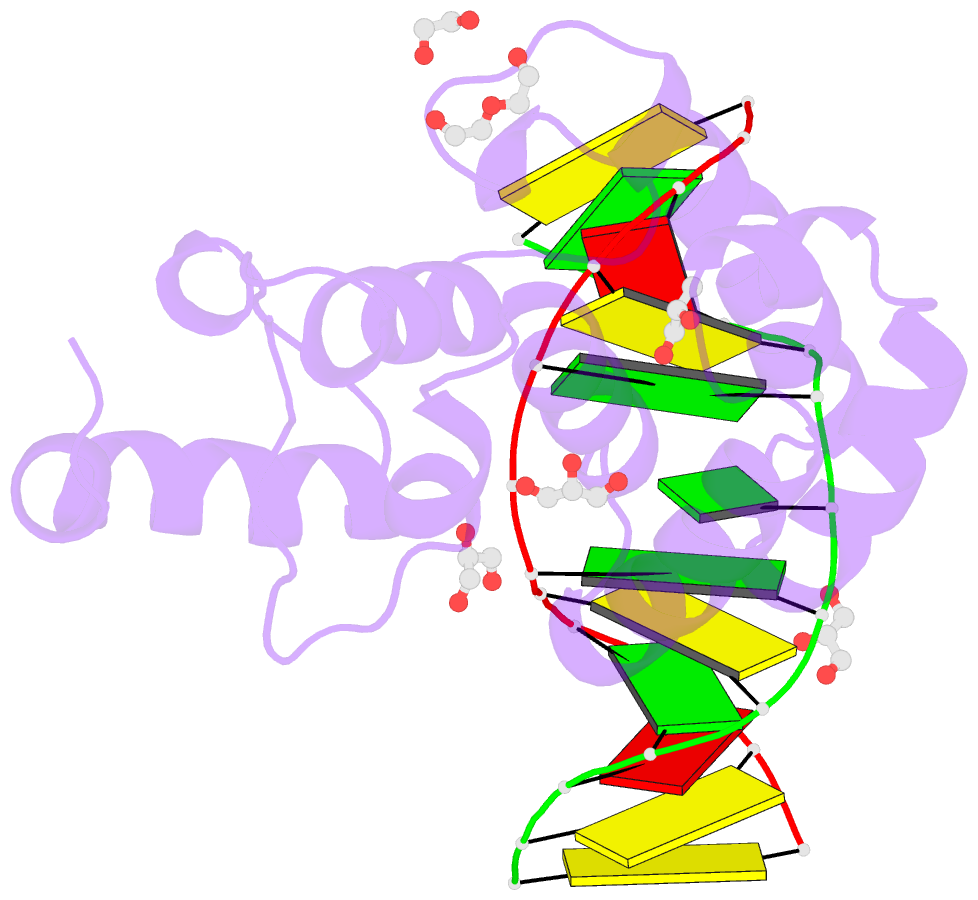
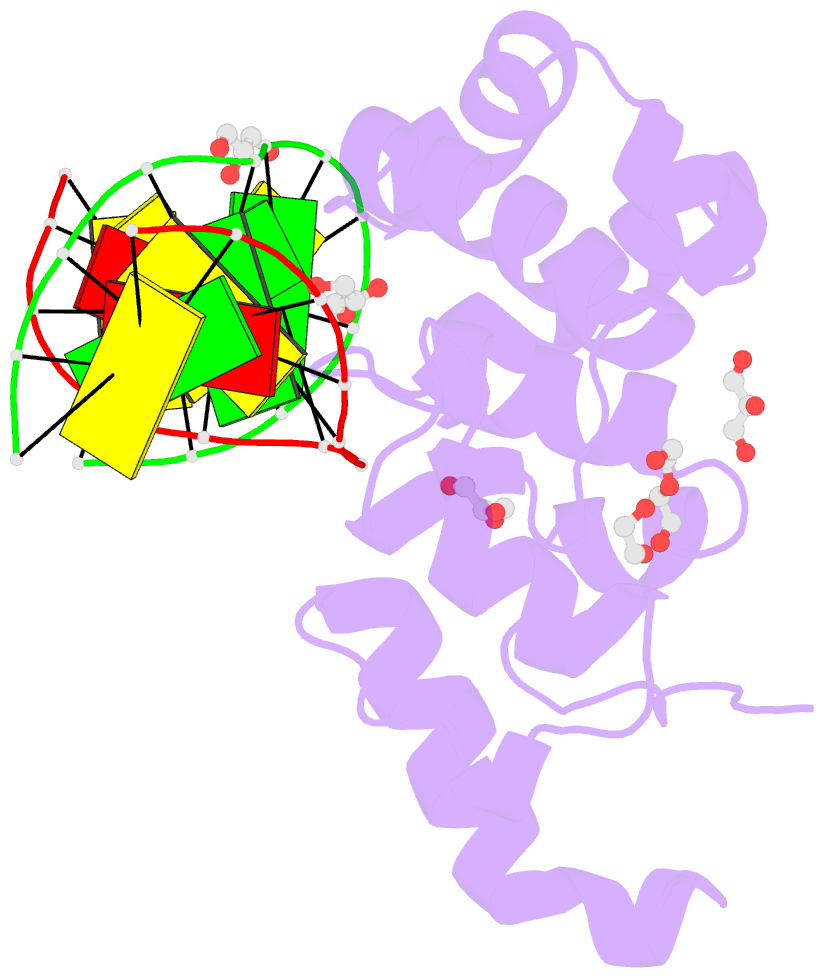
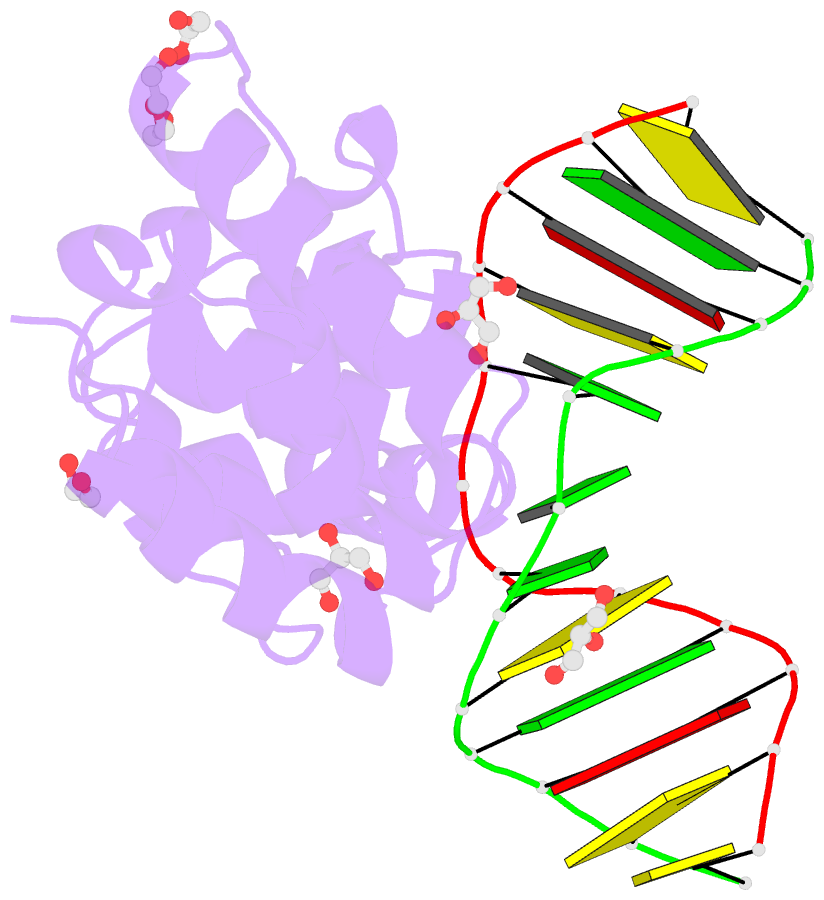
- Human mbd4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose
- Reference
- Pidugu LS, Bright H, Lin WJ, Majumdar C, Van Ostrand RP, David SS, Pozharski E, Drohat AC (2021): "Structural Insights into the Mechanism of Base Excision by MBD4." J.Mol.Biol., 433, 167097. doi: 10.1016/j.jmb.2021.167097.
- Abstract
- DNA glycosylases remove damaged or modified nucleobases by cleaving the N-glycosyl bond and the correct nucleotide is restored through subsequent base excision repair. In addition to excising threatening lesions, DNA glycosylases contribute to epigenetic regulation by mediating DNA demethylation and perform other important functions. However, the catalytic mechanism remains poorly defined for many glycosylases, including MBD4 (methyl-CpG binding domain IV), a member of the helix-hairpin-helix (HhH) superfamily. MBD4 excises thymine from G·T mispairs, suppressing mutations caused by deamination of 5-methylcytosine, and it removes uracil and modified uracils (e.g., 5-hydroxymethyluracil) mispaired with guanine. To investigate the mechanism of MBD4 we solved high-resolution structures of enzyme-DNA complexes at three stages of catalysis. Using a non-cleavable substrate analog, 2'-deoxy-pseudouridine, we determined the first structure of an enzyme-substrate complex for wild-type MBD4, which confirms interactions that mediate lesion recognition and suggests that a catalytic Asp, highly conserved in HhH enzymes, binds the putative nucleophilic water molecule and stabilizes the transition state. Observation that mutating the Asp (to Gly) reduces activity by 2700-fold indicates an important role in catalysis, but probably not one as the nucleophile in a double-displacement reaction, as previously suggested. Consistent with direct-displacement hydrolysis, a structure of the enzyme-product complex indicates a reaction leading to inversion of configuration. A structure with DNA containing 1-azadeoxyribose models a potential oxacarbenium-ion intermediate and suggests the Asp could facilitate migration of the electrophile towards the nucleophilic water. Finally, the structures provide detailed snapshots of the HhH motif, informing how these ubiquitous metal-binding elements mediate DNA binding.