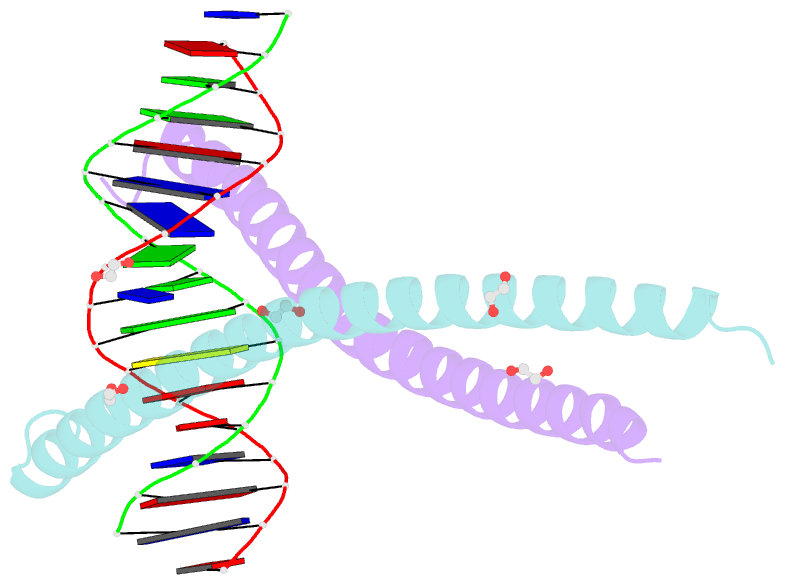
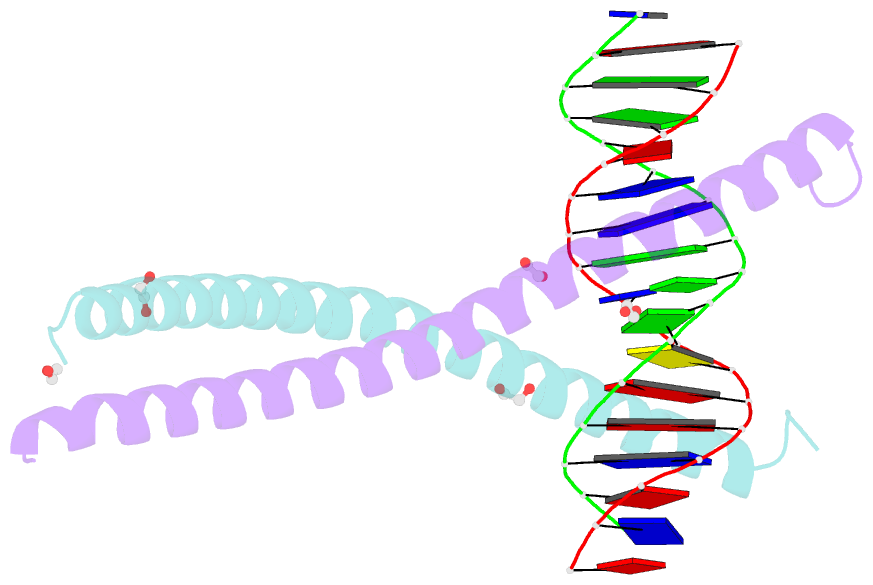
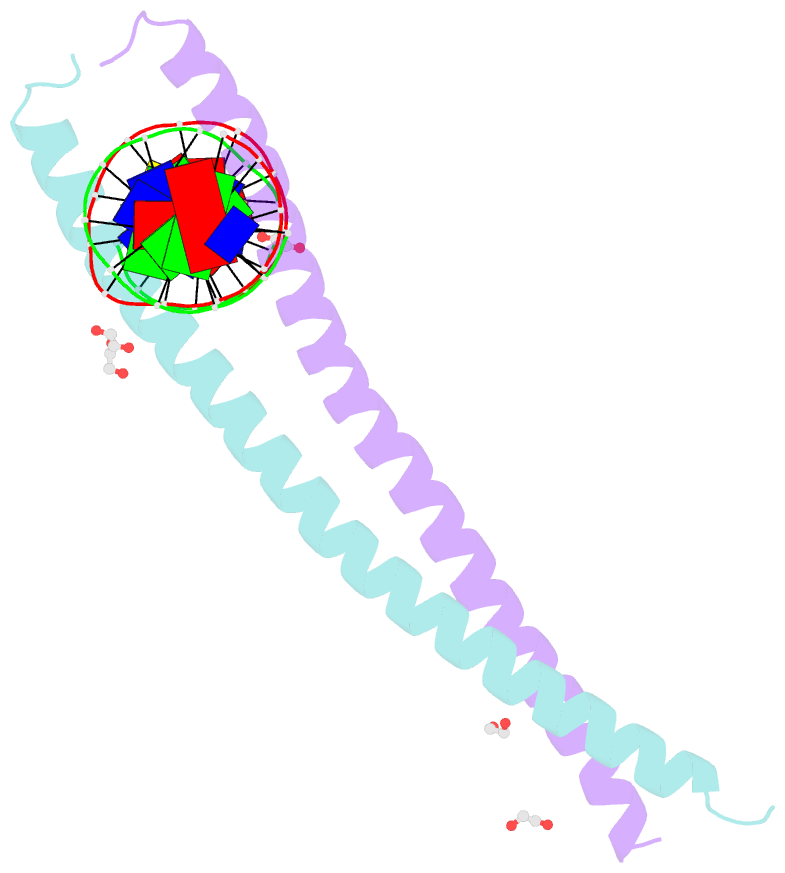
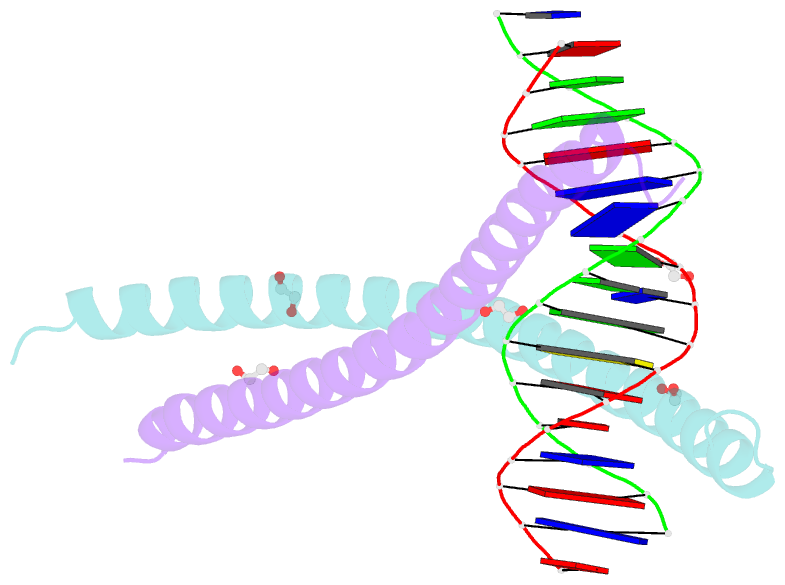
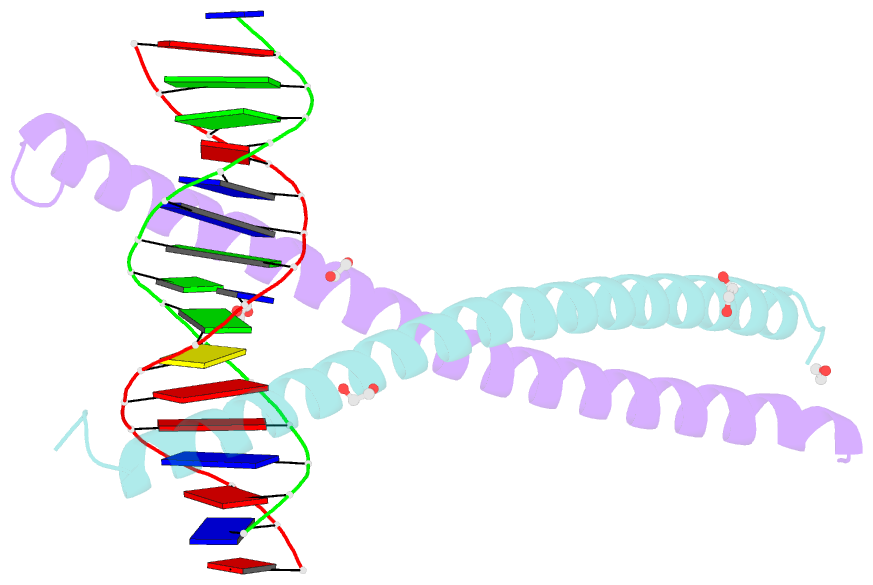
Summary information and primary citation
- PDB-id
- 7l4v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.75 Å)
- Summary
- C-terminal bzip domain of human c-ebpbeta bound to DNA with consensus recognition with gt mismatch
- Reference
- Yang J, Horton JR, Akdemir KC, Li J, Huang Y, Kumar J, Blumenthal RM, Zhang X, Cheng X (2021): "Preferential CEBP binding to T:G mismatches and increased C-to-T human somatic mutations." Nucleic Acids Res., 49, 5084-5094. doi: 10.1093/nar/gkab276.
- Abstract
- DNA cytosine methylation in mammals modulates gene expression and chromatin accessibility. It also impacts mutation rates, via spontaneous oxidative deamination of 5-methylcytosine (5mC) to thymine. In most cases the resulting T:G mismatches are repaired, following T excision by one of the thymine DNA glycosylases, TDG or MBD4. We found that C-to-T mutations are enriched in the binding sites of CCAAT/enhancer binding proteins (CEBP). Within a CEBP site, the presence of a T:G mismatch increased CEBPβ binding affinity by a factor of >60 relative to the normal C:G base pair. This enhanced binding to a mismatch inhibits its repair by both TDG and MBD4 in vitro. Furthermore, repair of the deamination product of unmethylated cytosine, which yields a U:G DNA mismatch that is normally repaired via uracil DNA glycosylase, is also inhibited by CEBPβ binding. Passage of a replication fork over either a T:G or U:G mismatch, before repair can occur, results in a C-to-T mutation in one of the daughter duplexes. Our study thus provides a plausible mechanism for accumulation of C-to-T human somatic mutations.