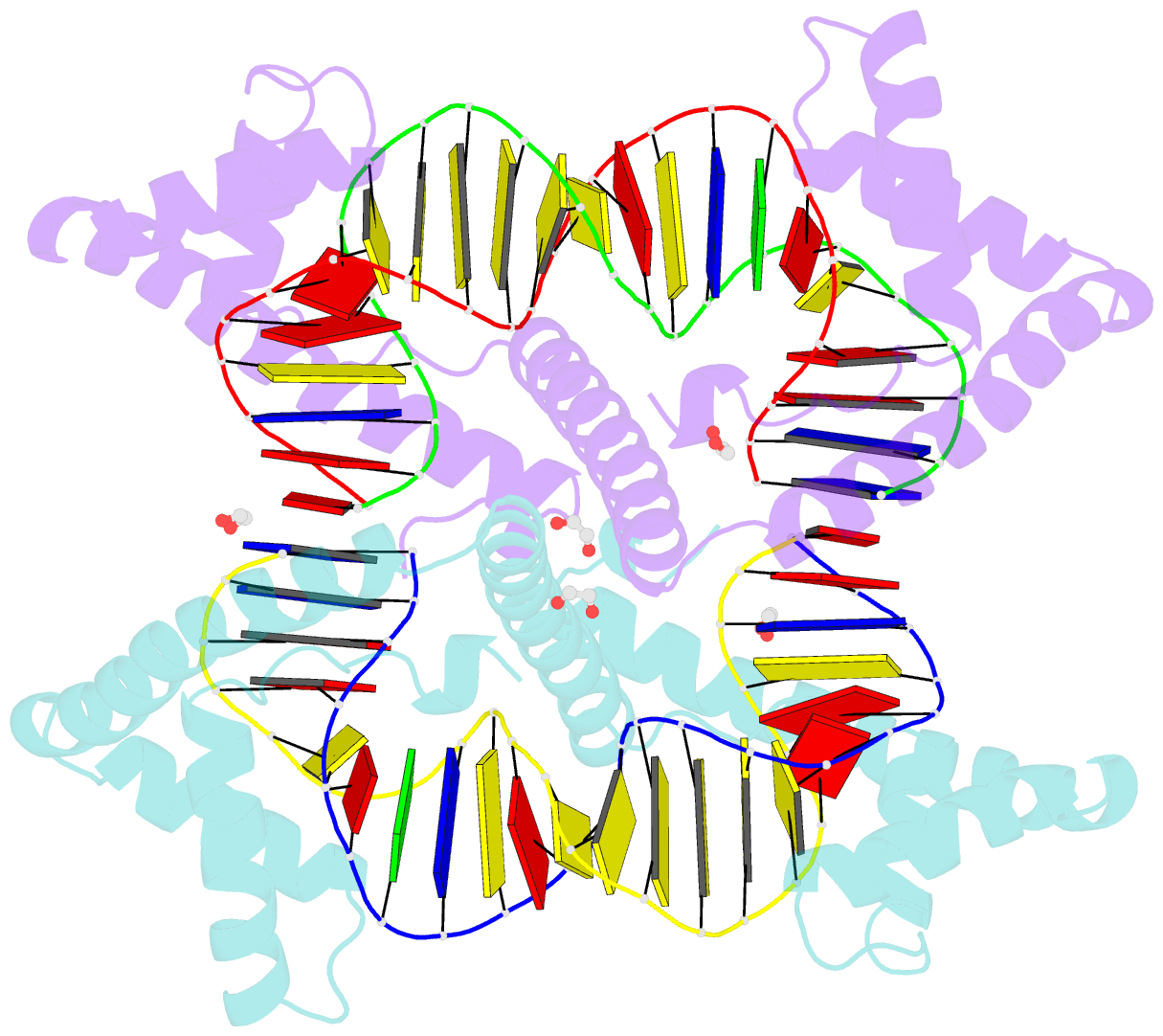
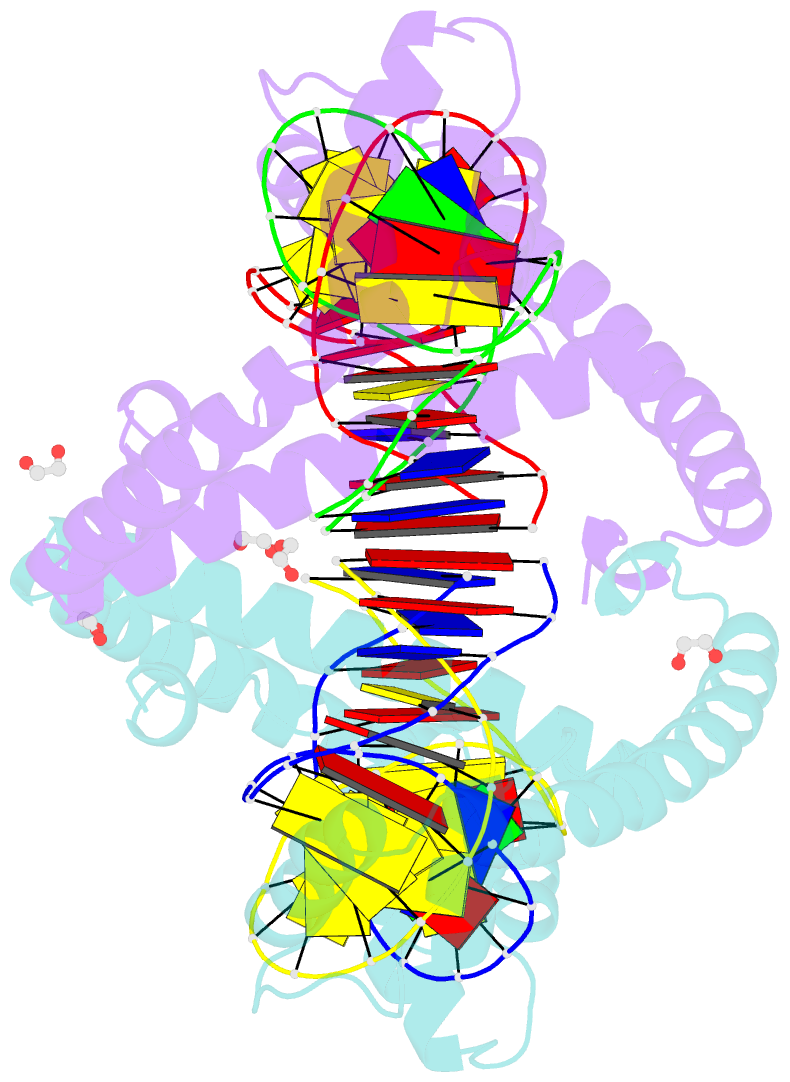
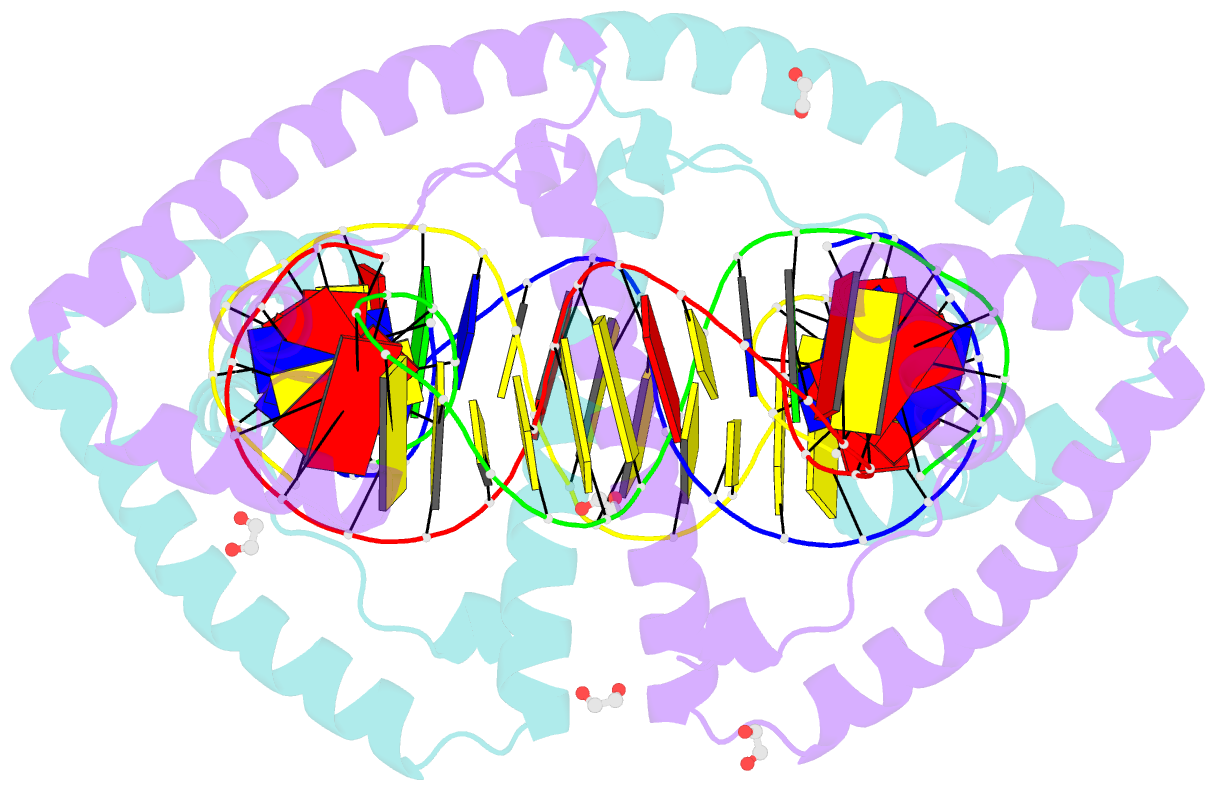
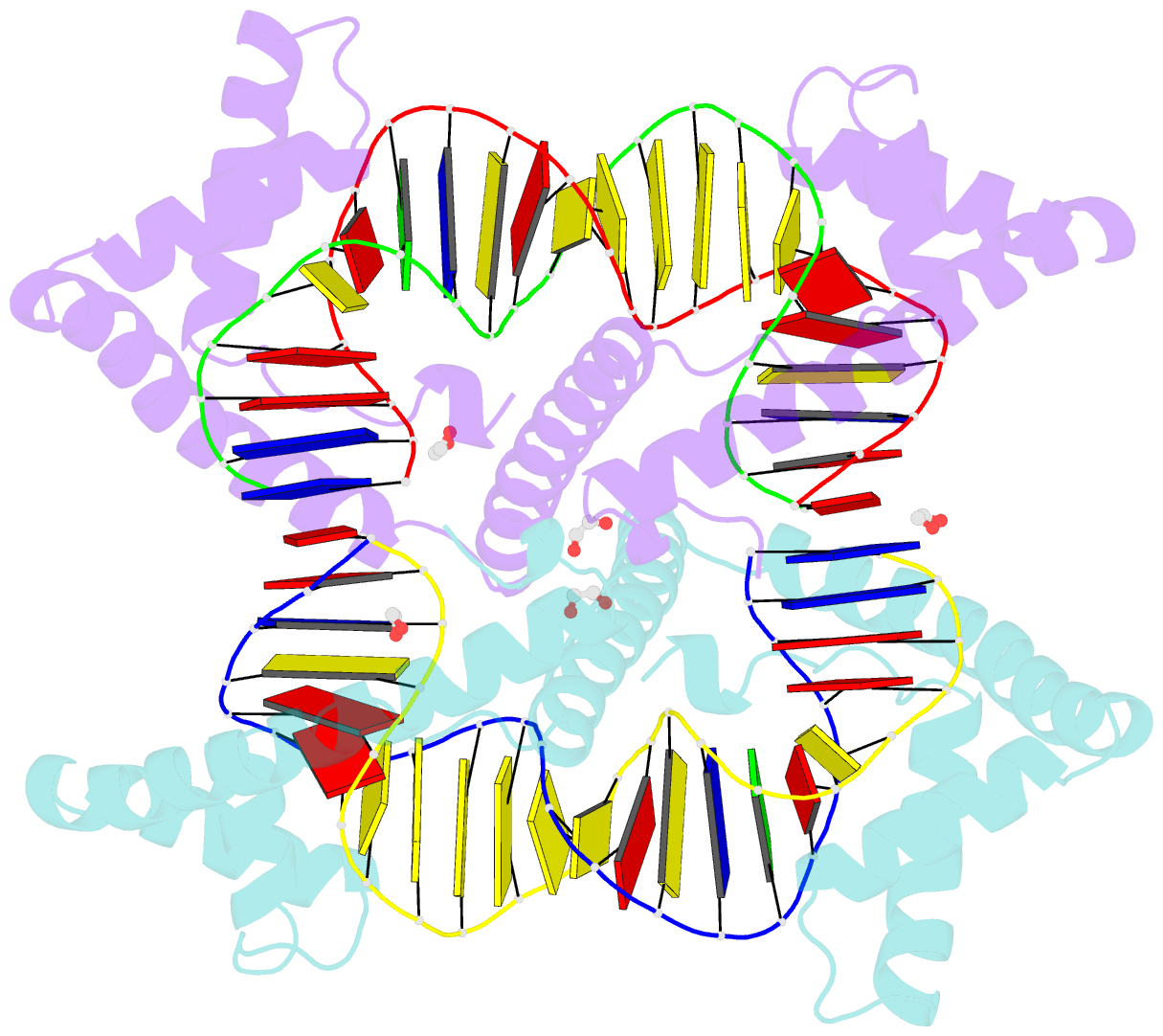
Summary information and primary citation
- PDB-id
- 7lbx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.7 Å)
- Summary
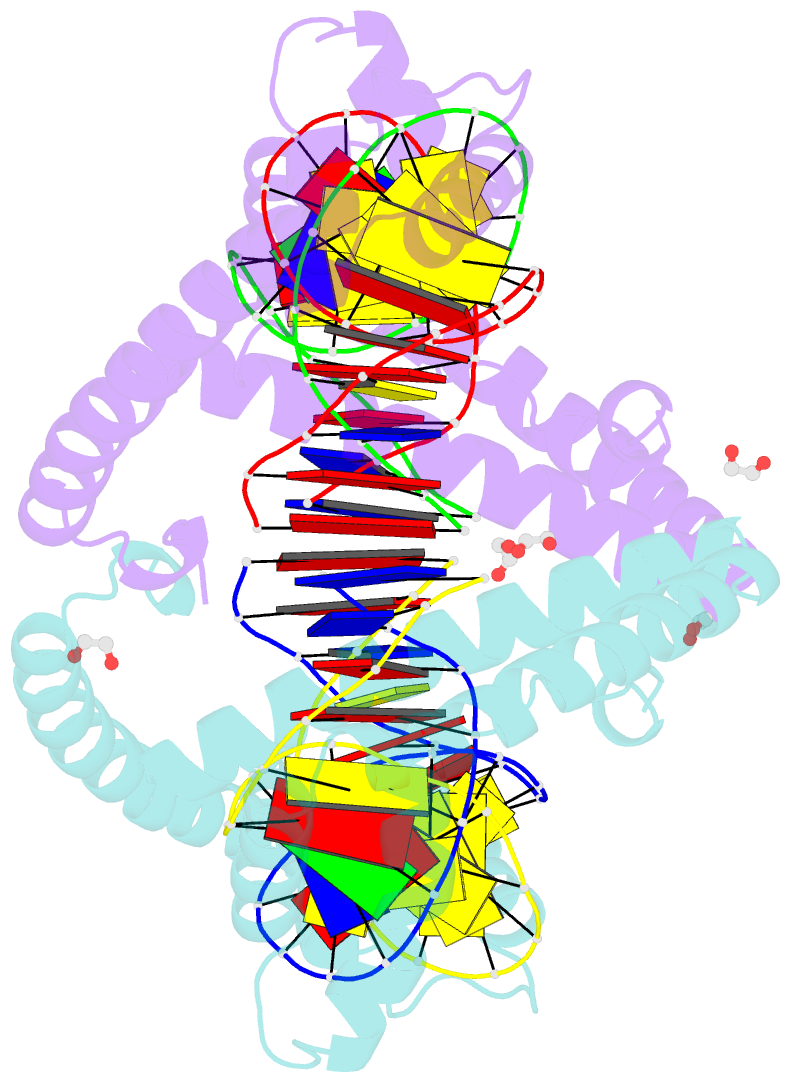
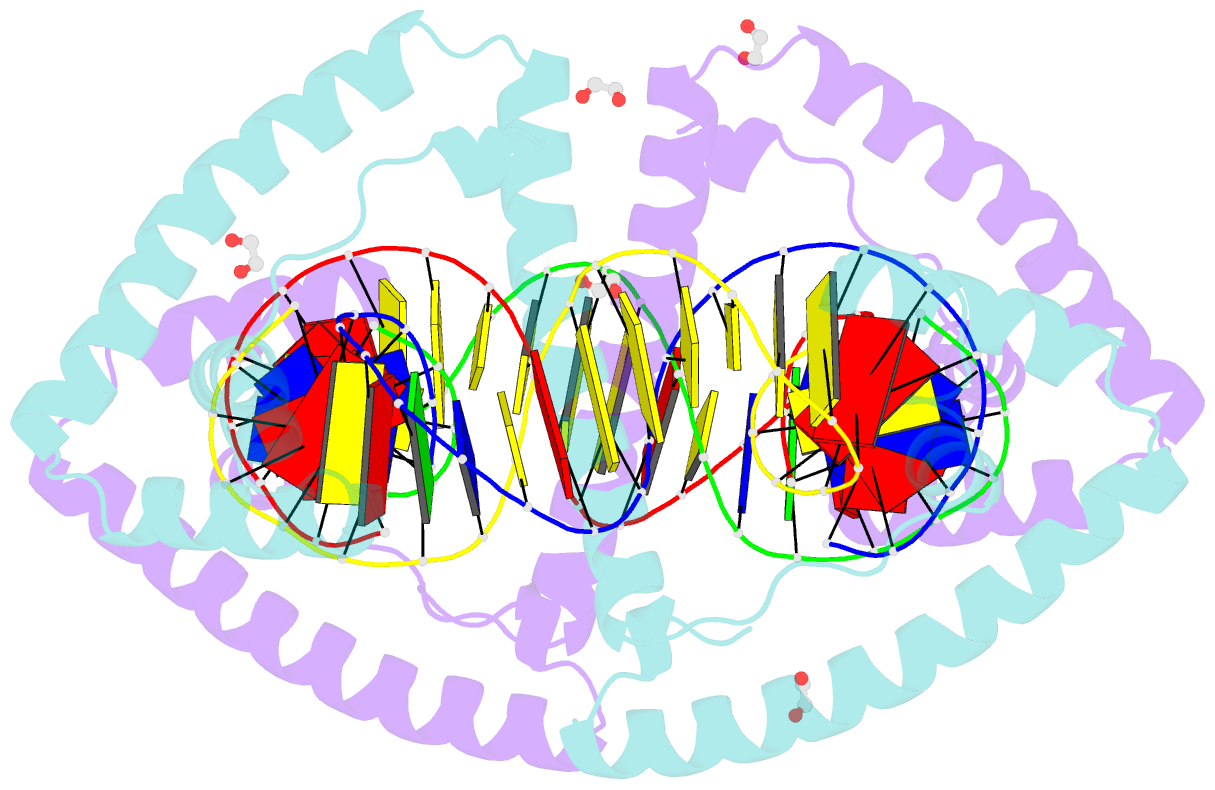
- Crystal structure of tfam (mitochondrial transcription factor a) in complex with lsp
- Reference
- Choi WS, Garcia-Diaz M (2022): "A minimal motif for sequence recognition by mitochondrial transcription factor A (TFAM)." Nucleic Acids Res., 50, 322-332. doi: 10.1093/nar/gkab1230.
- Abstract
- Mitochondrial transcription factor A (TFAM) plays a critical role in mitochondrial transcription initiation and mitochondrial DNA (mtDNA) packaging. Both functions require DNA binding, but in one case TFAM must recognize a specific promoter sequence, while packaging requires coating of mtDNA by association with non sequence-specific regions. The mechanisms by which TFAM achieves both sequence-specific and non sequence-specific recognition have not yet been determined. Existing crystal structures of TFAM bound to DNA allowed us to identify two guanine-specific interactions that are established between TFAM and the bound DNA. These interactions are observed when TFAM is bound to both specific promoter sequences and non-sequence specific DNA. These interactions are established with two guanine bases separated by 10 random nucleotides (GN10G). Our biochemical results demonstrate that the GN10G consensus is essential for transcriptional initiation and contributes to facilitating TFAM binding to DNA substrates. Furthermore, we report a crystal structure of TFAM in complex with a non sequence-specific sequence containing a GN10G consensus. The structure reveals a unique arrangement in which TFAM bridges two DNA substrates while maintaining the GN10G interactions. We propose that the GN10G consensus is key to facilitate the interaction of TFAM with DNA.