Summary information and primary citation
- PDB-id
- 7lcc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination
- Method
- cryo-EM (3.66 Å)
- Summary
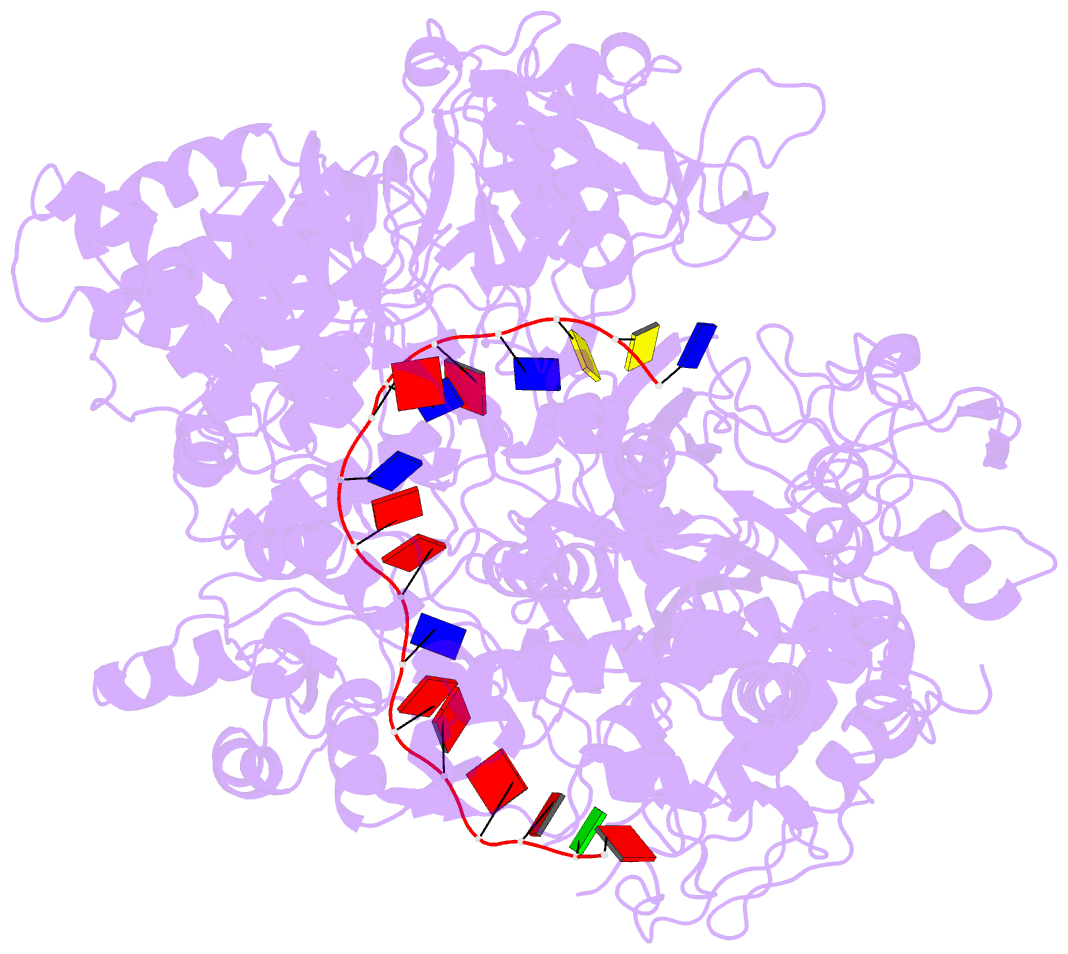
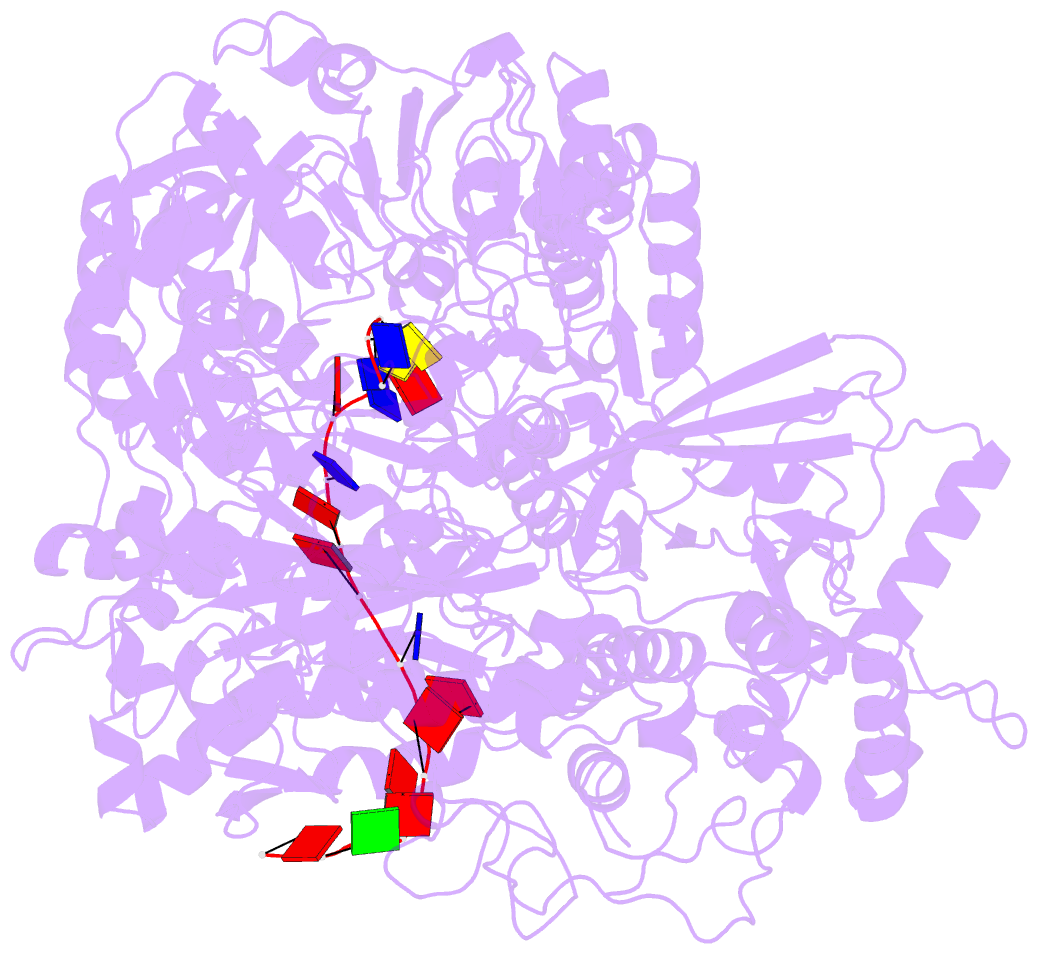
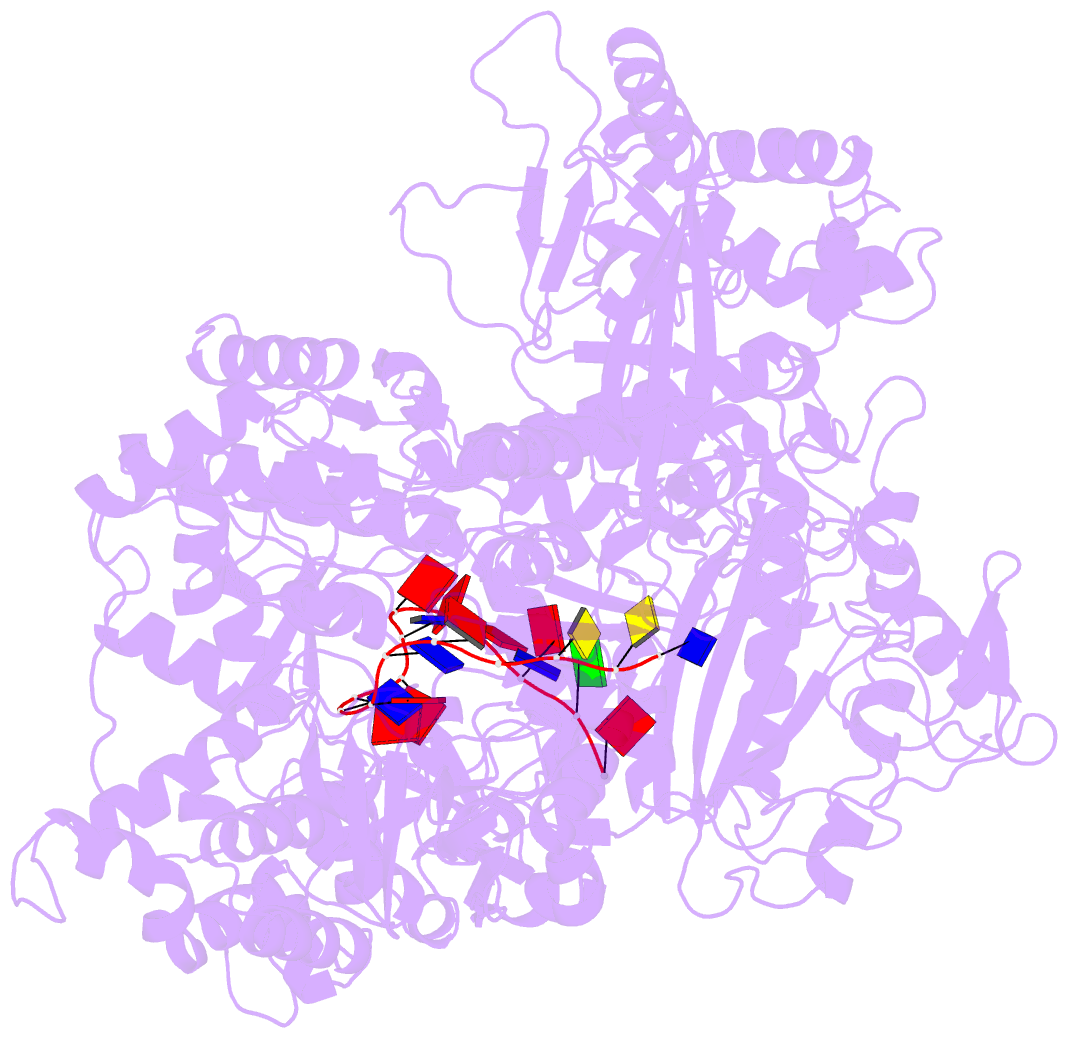
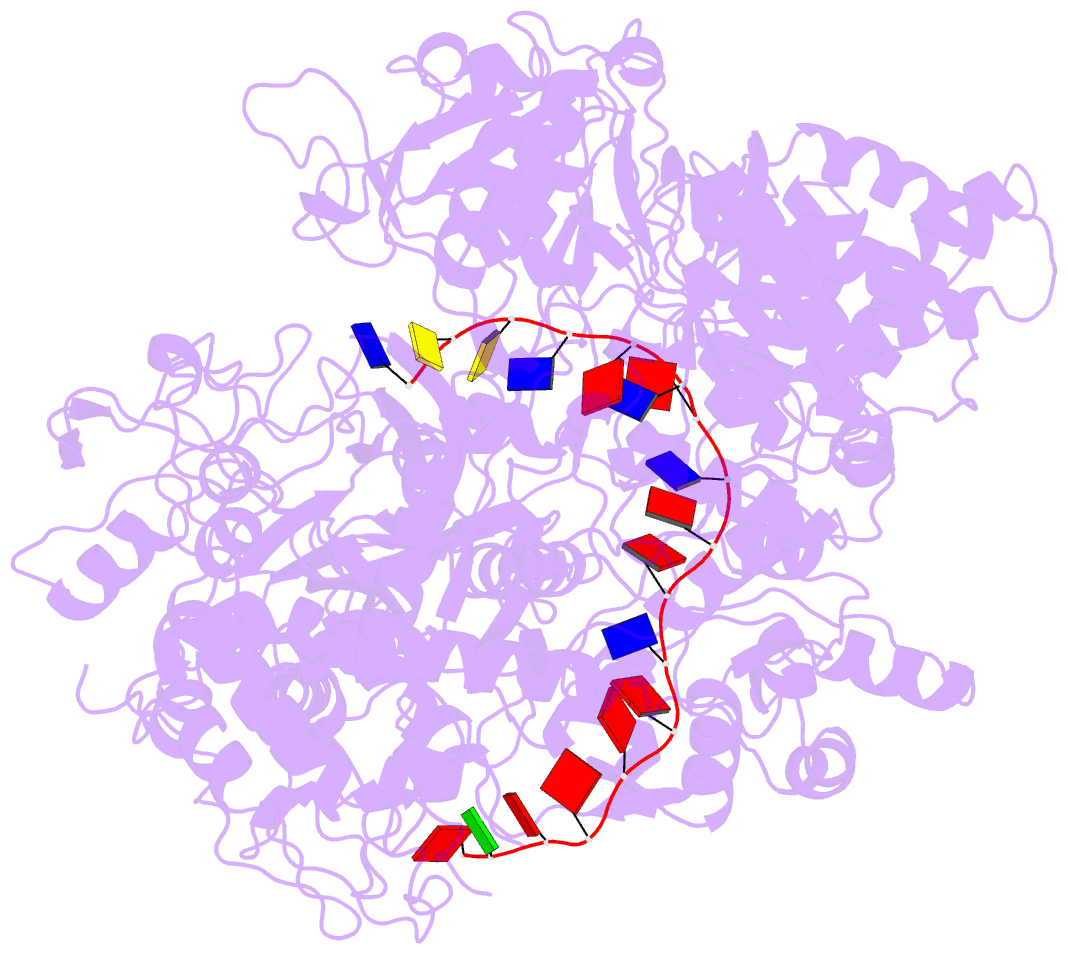
- Helitron transposase bound to lts
- Reference
- Kosek D, Grabundzija I, Lei H, Bilic I, Wang H, Jin Y, Peaslee GF, Hickman AB, Dyda F (2021): "The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end." Mol.Cell, 81, 4271-4286.e4. doi: 10.1016/j.molcel.2021.07.028.
- Abstract
- Helitrons are widespread eukaryotic DNA transposons that have significantly contributed to genome variability and evolution, in part because of their distinctive, replicative rolling-circle mechanism, which often mobilizes adjacent genes. Although most eukaryotic transposases form oligomers and use RNase H-like domains to break and rejoin double-stranded DNA (dsDNA), Helitron transposases contain a single-stranded DNA (ssDNA)-specific HUH endonuclease domain. Here, we report the cryo-electron microscopy structure of a Helitron transposase bound to the 5'-transposon end, providing insight into its multidomain architecture and function. The monomeric transposase forms a tightly packed assembly that buries the covalently attached cleaved end, protecting it until the second end becomes available. The structure reveals unexpected architectural similarity to TraI, a bacterial relaxase that also catalyzes ssDNA movement. The HUH active site suggests how two juxtaposed tyrosines, a feature of many replication initiators that use HUH nucleases, couple the conformational shift of an α-helix to control strand cleavage and ligation reactions.