Summary information and primary citation
- PDB-id
- 7lnj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.68 Å)
- Summary
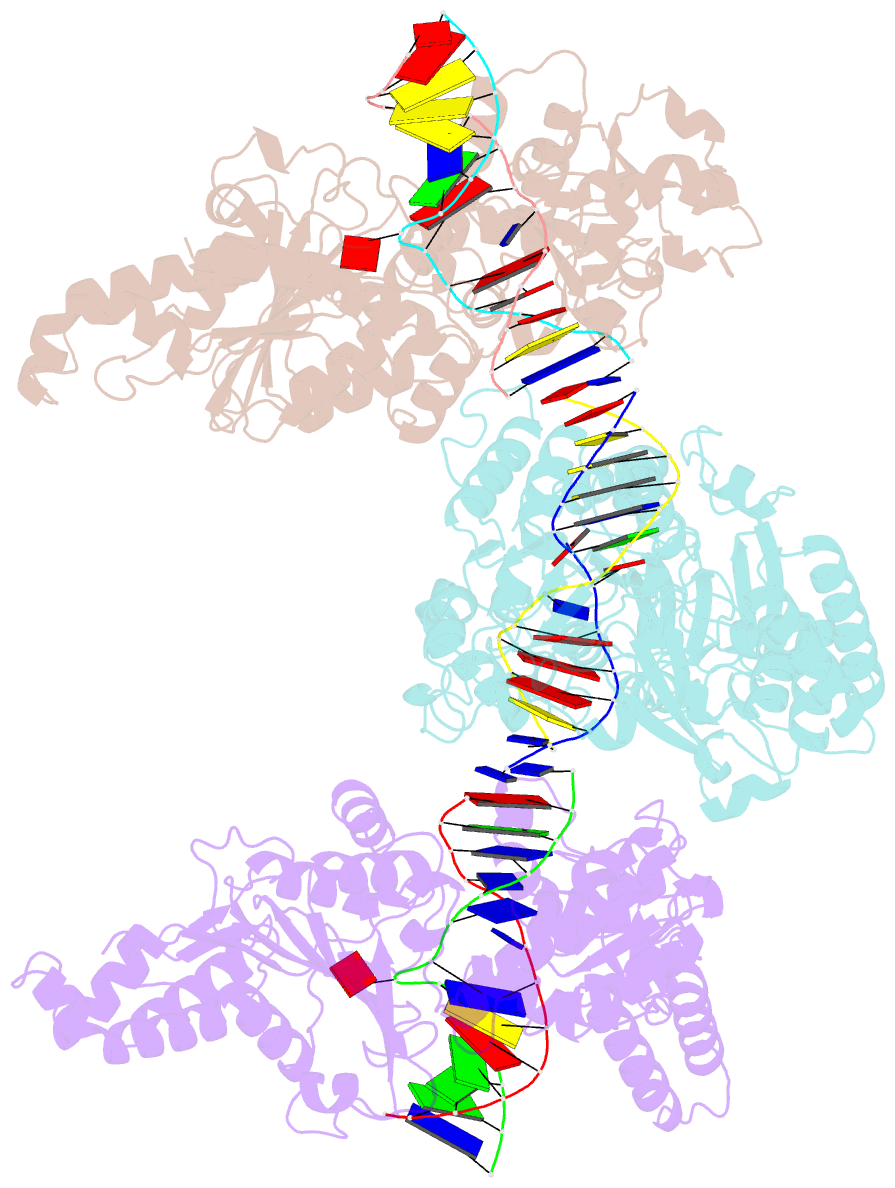
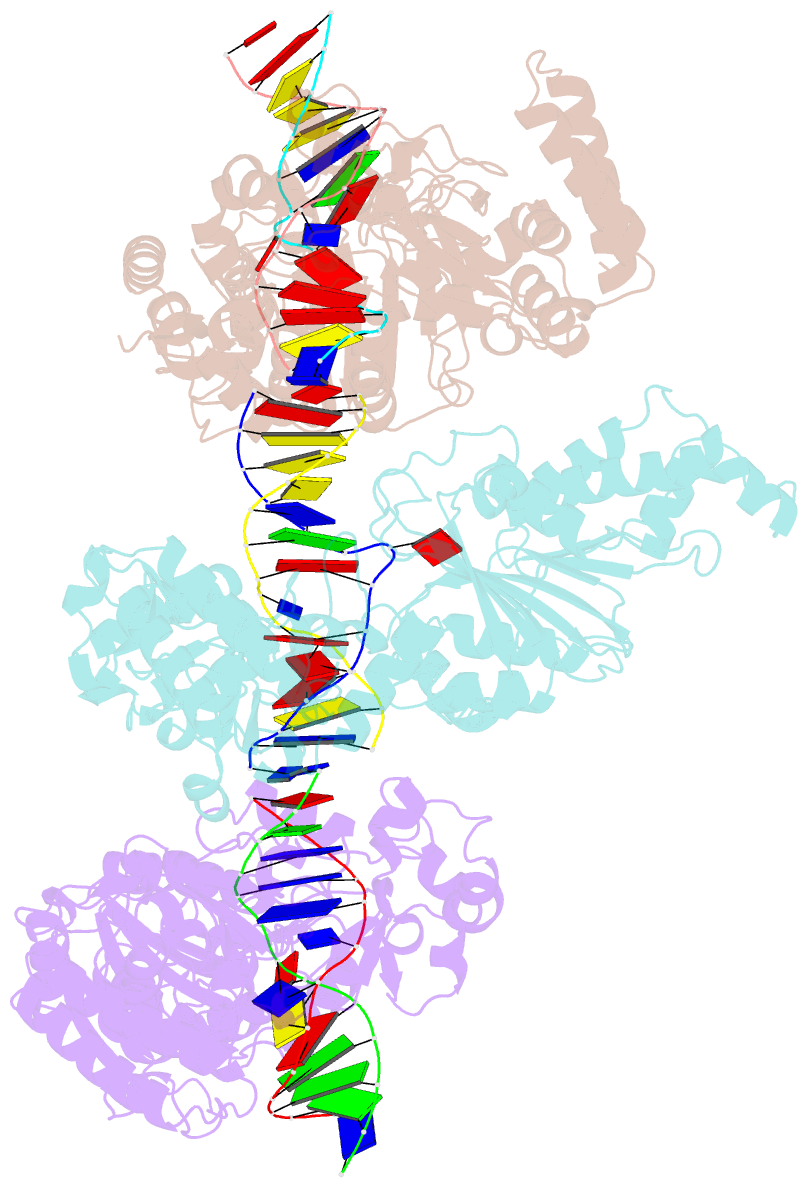
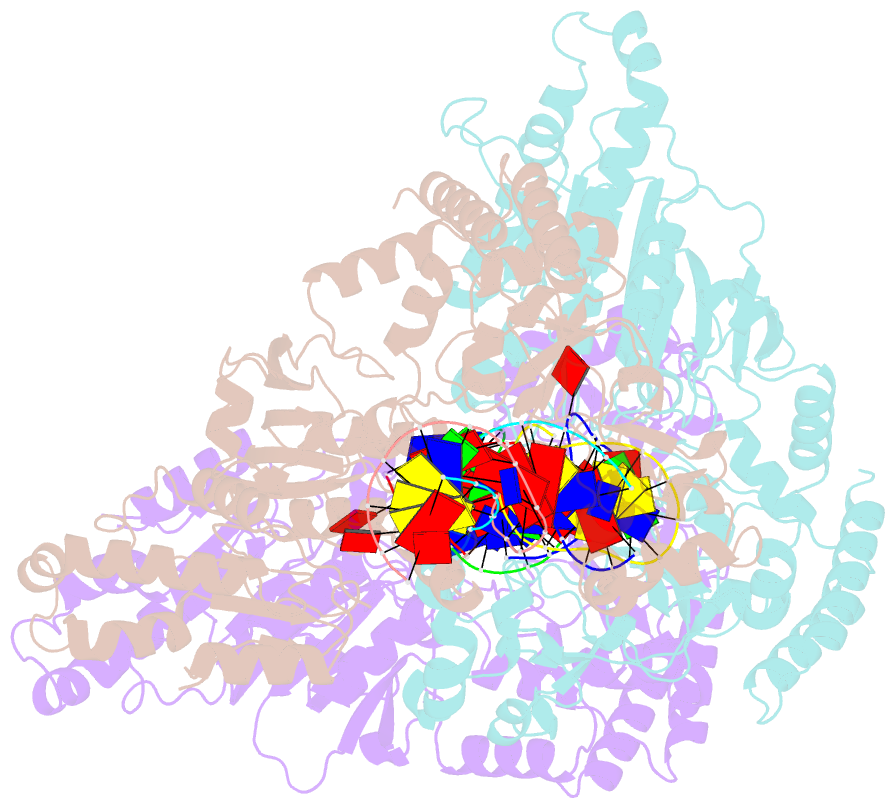
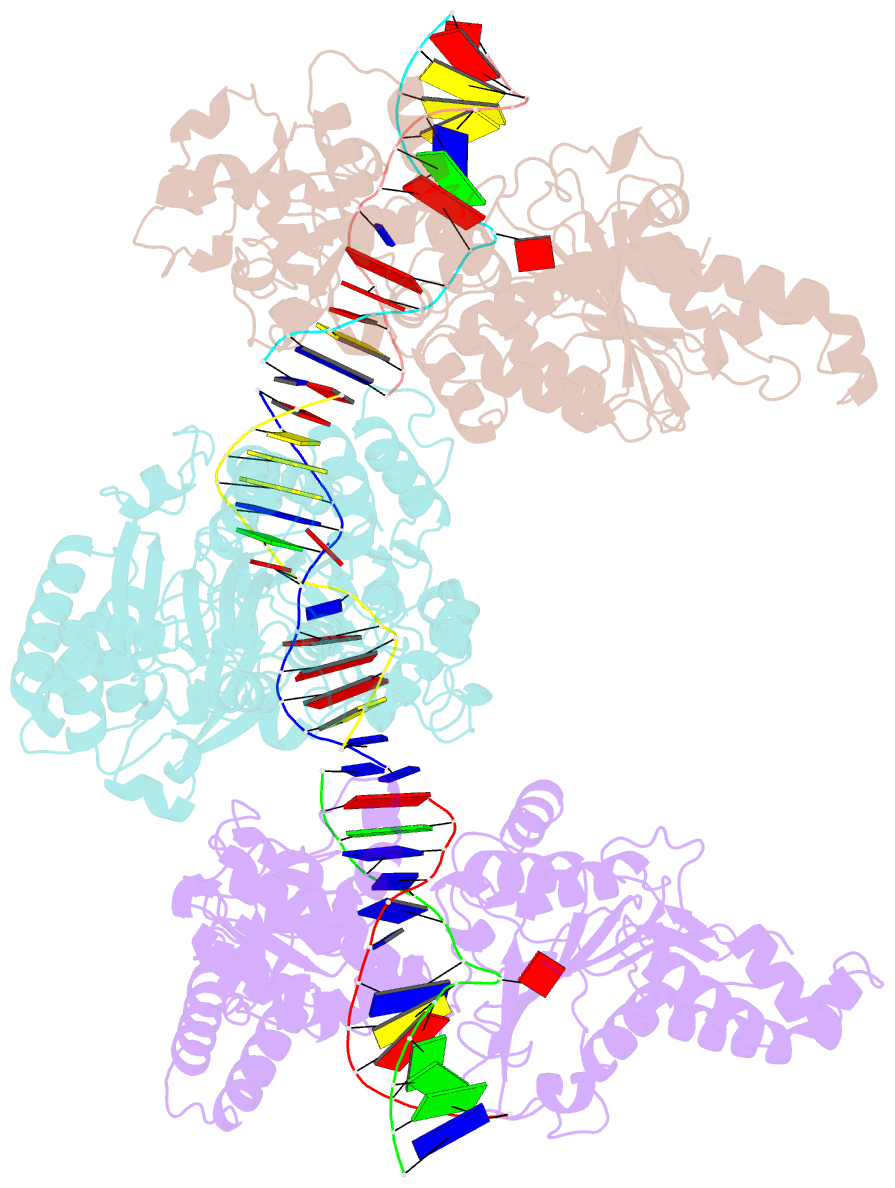
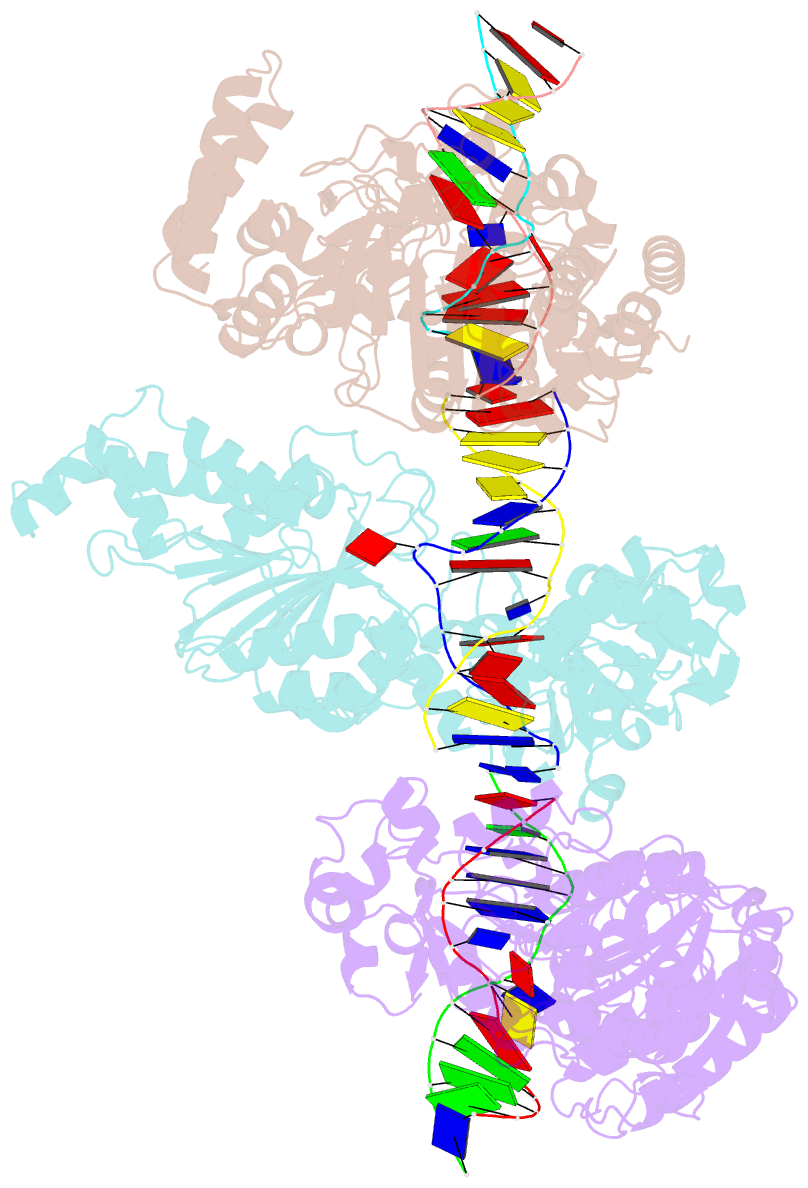
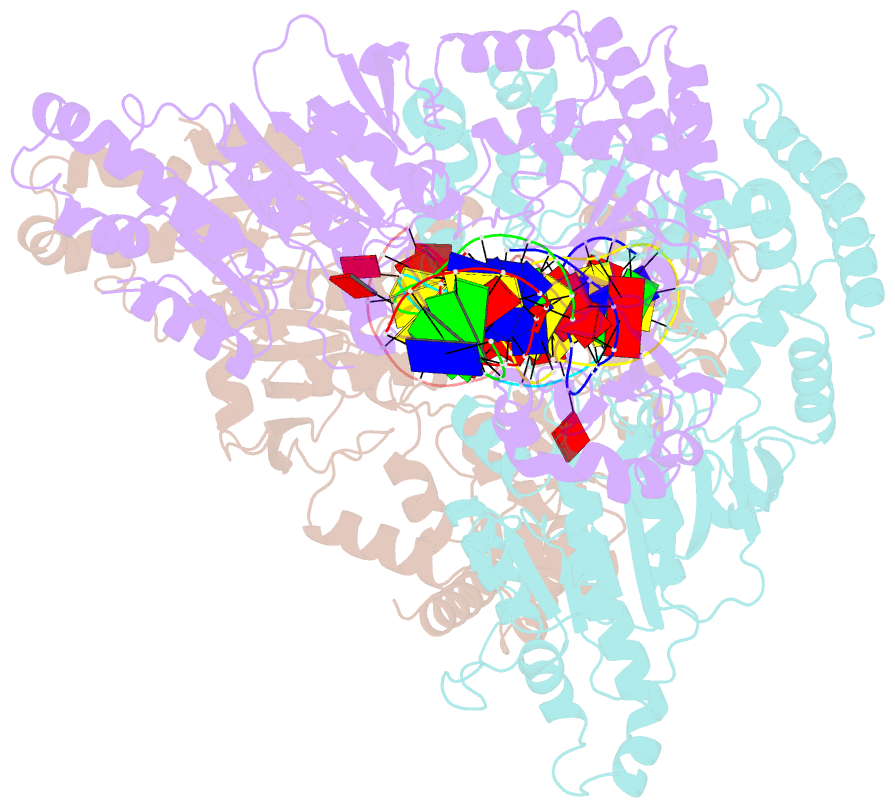
- Cama adenine methyltransferase complexed to cognate substrate DNA
- Reference
- Zhou J, Horton JR, Blumenthal RM, Zhang X, Cheng X (2021): "Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix." Nat Commun, 12, 3436. doi: 10.1038/s41467-021-23693-w.
- Abstract
- Clostridioides difficile infections are an urgent medical problem. The newly discovered C. difficile adenine methyltransferase A (CamA) is specified by all C. difficile genomes sequenced to date (>300), but is rare among other bacteria. CamA is an orphan methyltransferase, unassociated with a restriction endonuclease. CamA-mediated methylation at CAAAAA is required for normal sporulation, biofilm formation, and intestinal colonization by C. difficile. We characterized CamA kinetic parameters, and determined its structure bound to DNA containing the recognition sequence. CamA contains an N-terminal domain for catalyzing methyl transfer, and a C-terminal DNA recognition domain. Major and minor groove DNA contacts in the recognition site involve base-specific hydrogen bonds, van der Waals contacts and the Watson-Crick pairing of a rearranged A:T base pair. These provide sufficient sequence discrimination to ensure high specificity. Finally, the surprisingly weak binding of the methyl donor S-adenosyl-L-methionine (SAM) might provide avenues for inhibiting CamA activity using SAM analogs.