Summary information and primary citation
- PDB-id
- 7lt3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (4.6 Å)
- Summary
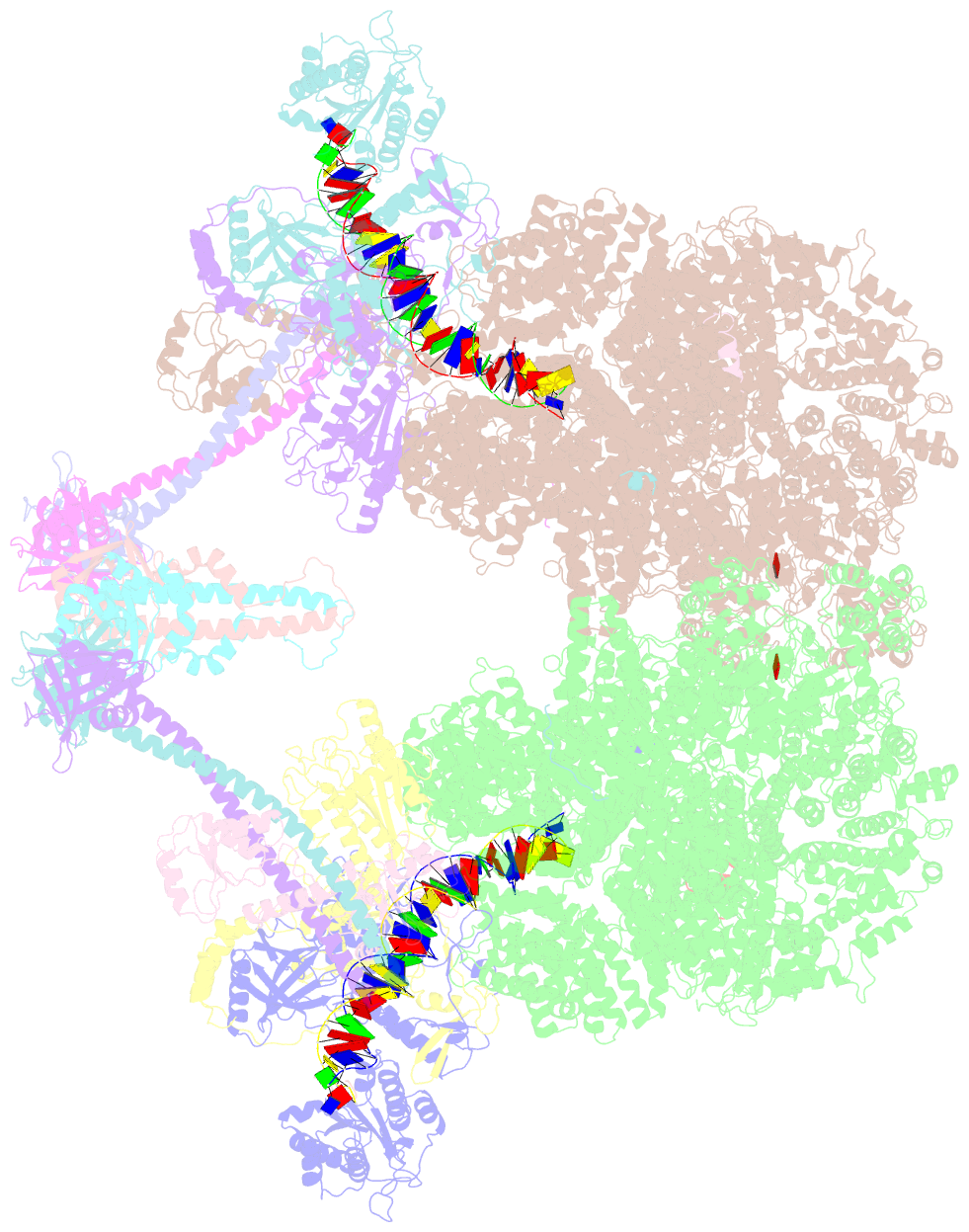
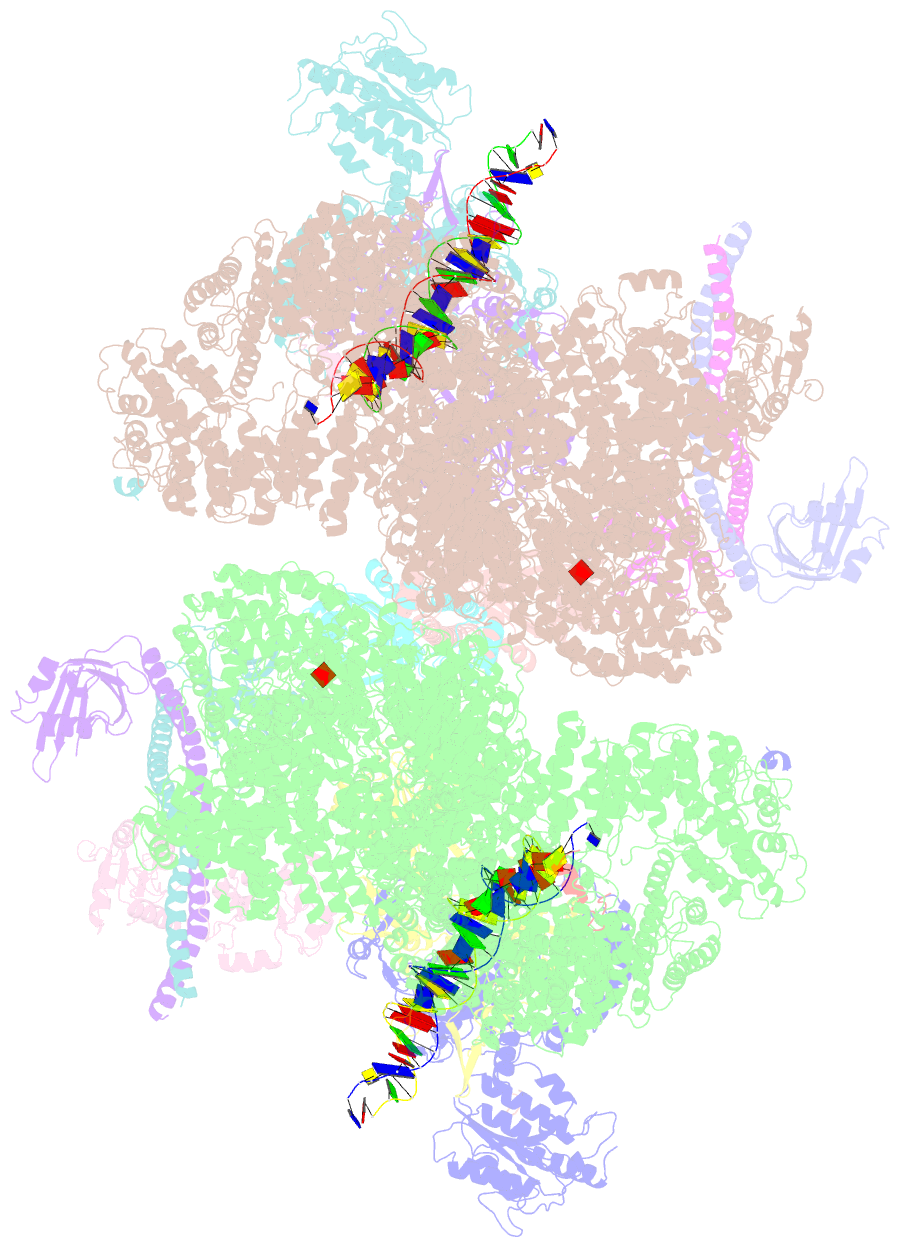
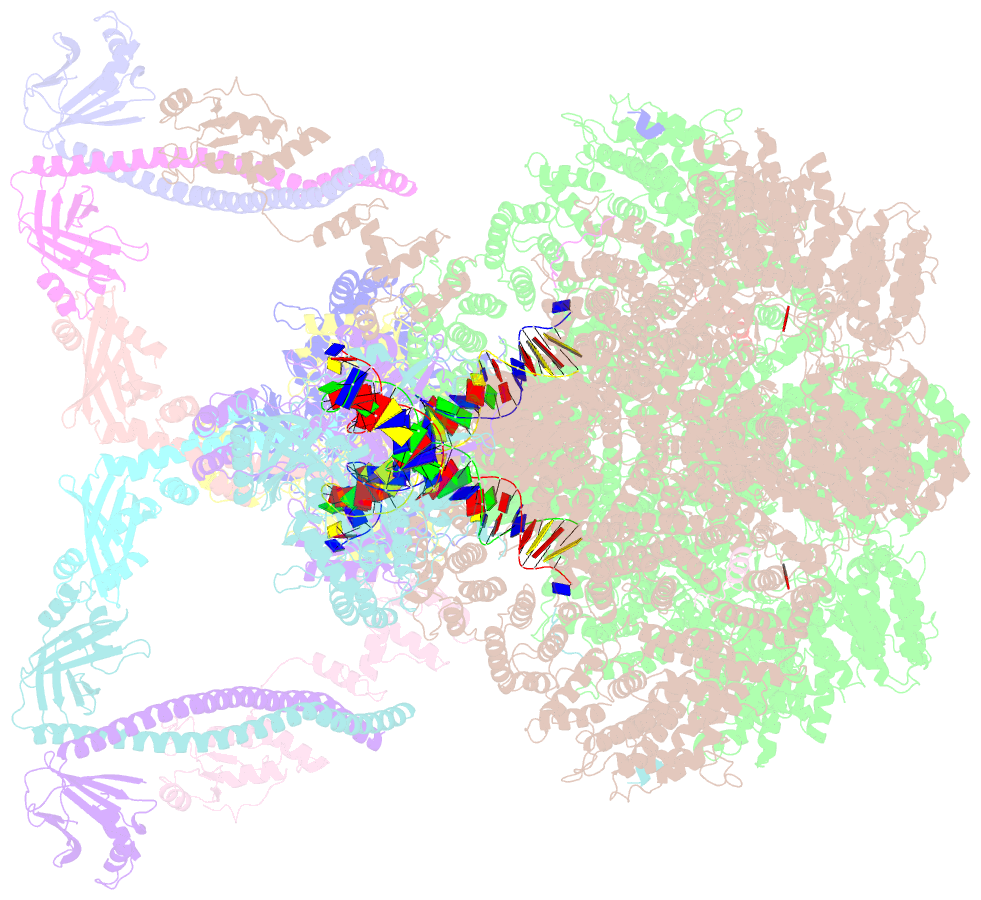
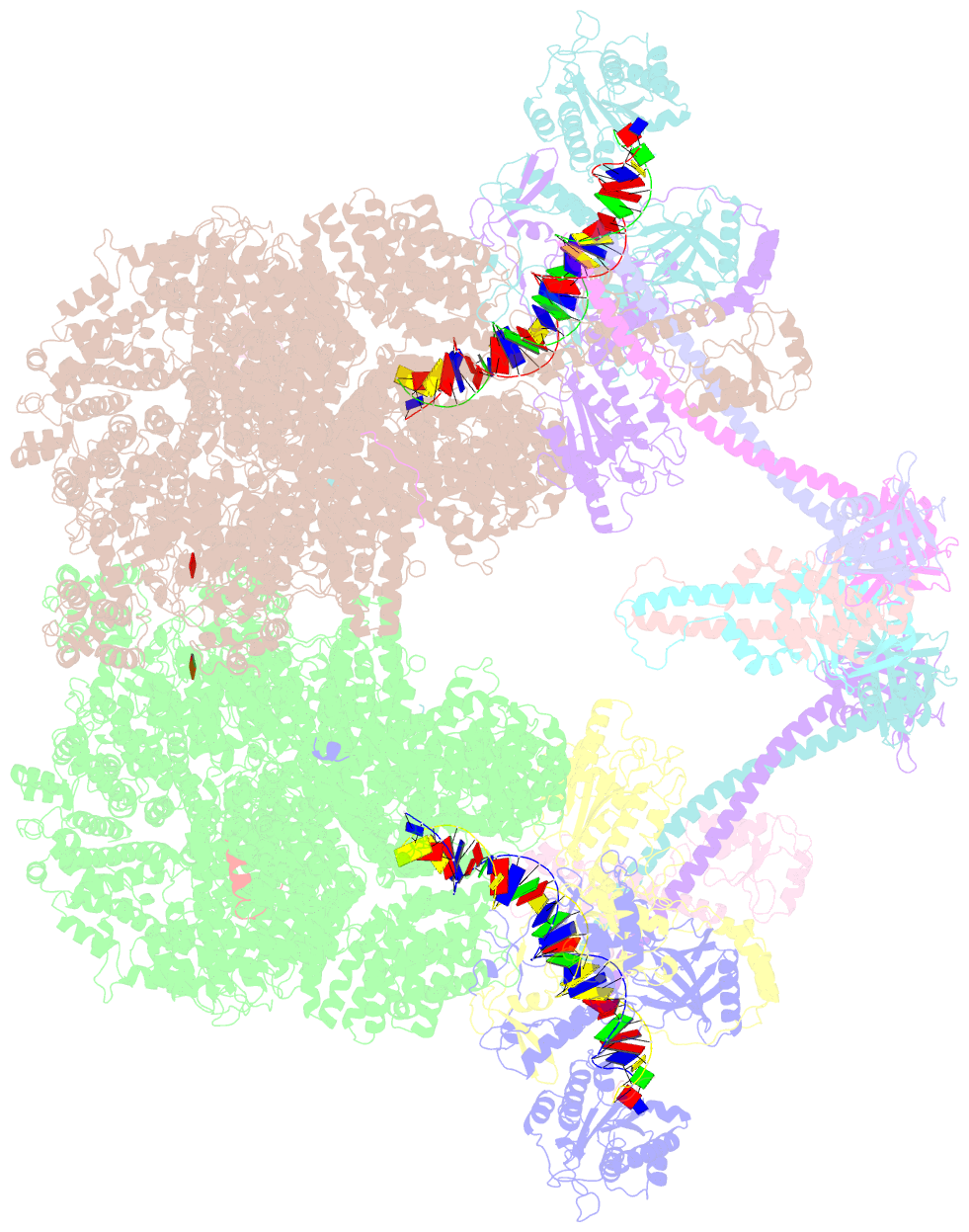
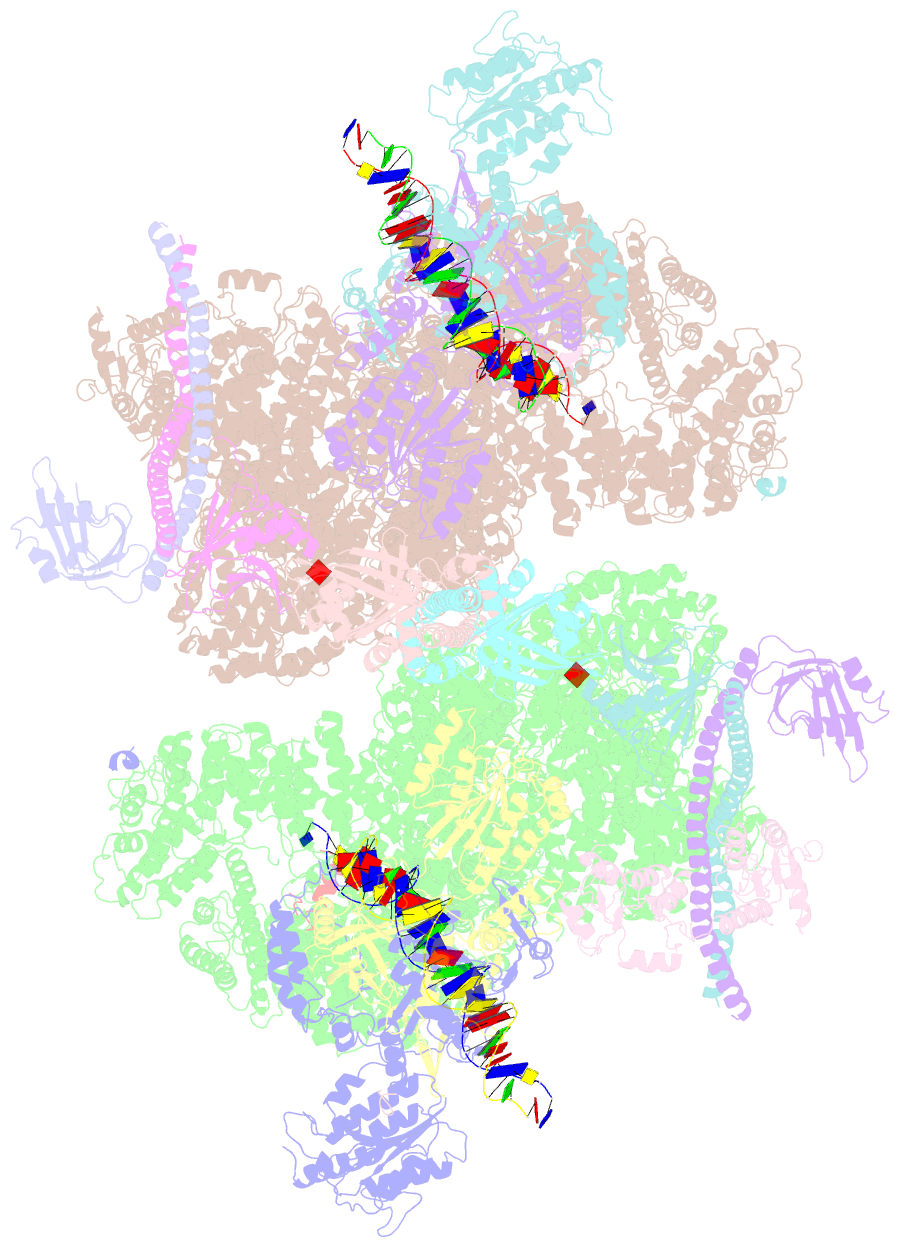
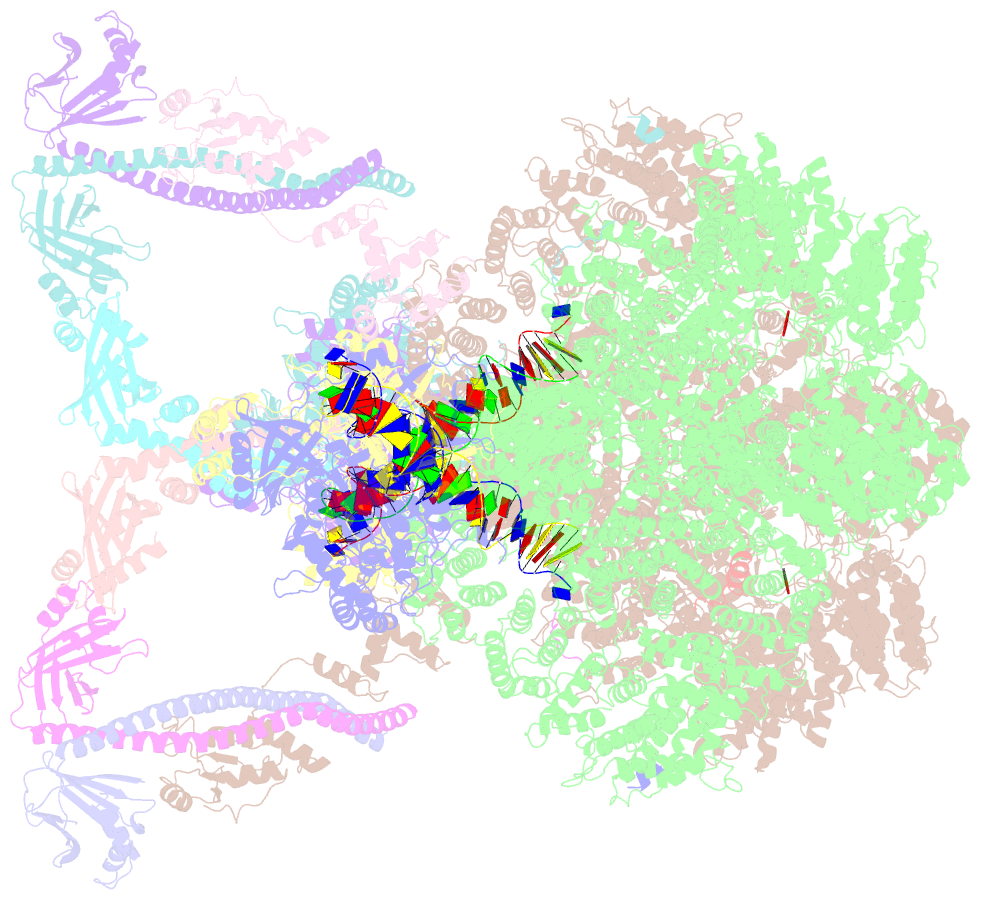
- Nhej long-range synaptic complex
- Reference
- Chen S, Lee L, Naila T, Fishbain S, Wang A, Tomkinson AE, Lees-Miller SP, He Y (2021): "Structural basis of long-range to short-range synaptic transition in NHEJ." Nature, 593, 294-298. doi: 10.1038/s41586-021-03458-7.
- Abstract
- DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis1,2. The conserved error-prone non-homologous end joining (NHEJ) pathway has a key role in determining the effects of DSB-inducing agents that are used to treat cancer as well as the generation of the diversity in antibodies and T cell receptors2,3. Here we applied single-particle cryo-electron microscopy to visualize two key DNA-protein complexes that are formed by human NHEJ factors. The Ku70/80 heterodimer (Ku), the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), DNA ligase IV (LigIV), XRCC4 and XLF form a long-range synaptic complex, in which the DNA ends are held approximately 115 Å apart. Two DNA end-bound subcomplexes comprising Ku and DNA-PKcs are linked by interactions between the DNA-PKcs subunits and a scaffold comprising LigIV, XRCC4, XLF, XRCC4 and LigIV. The relative orientation of the DNA-PKcs molecules suggests a mechanism for autophosphorylation in trans, which leads to the dissociation of DNA-PKcs and the transition into the short-range synaptic complex. Within this complex, the Ku-bound DNA ends are aligned for processing and ligation by the XLF-anchored scaffold, and a single catalytic domain of LigIV is stably associated with a nick between the two Ku molecules, which suggests that the joining of both strands of a DSB involves both LigIV molecules.