Summary information and primary citation
- PDB-id
- 7lxh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA-antibiotic
- Method
- X-ray (1.667 Å)
- Summary
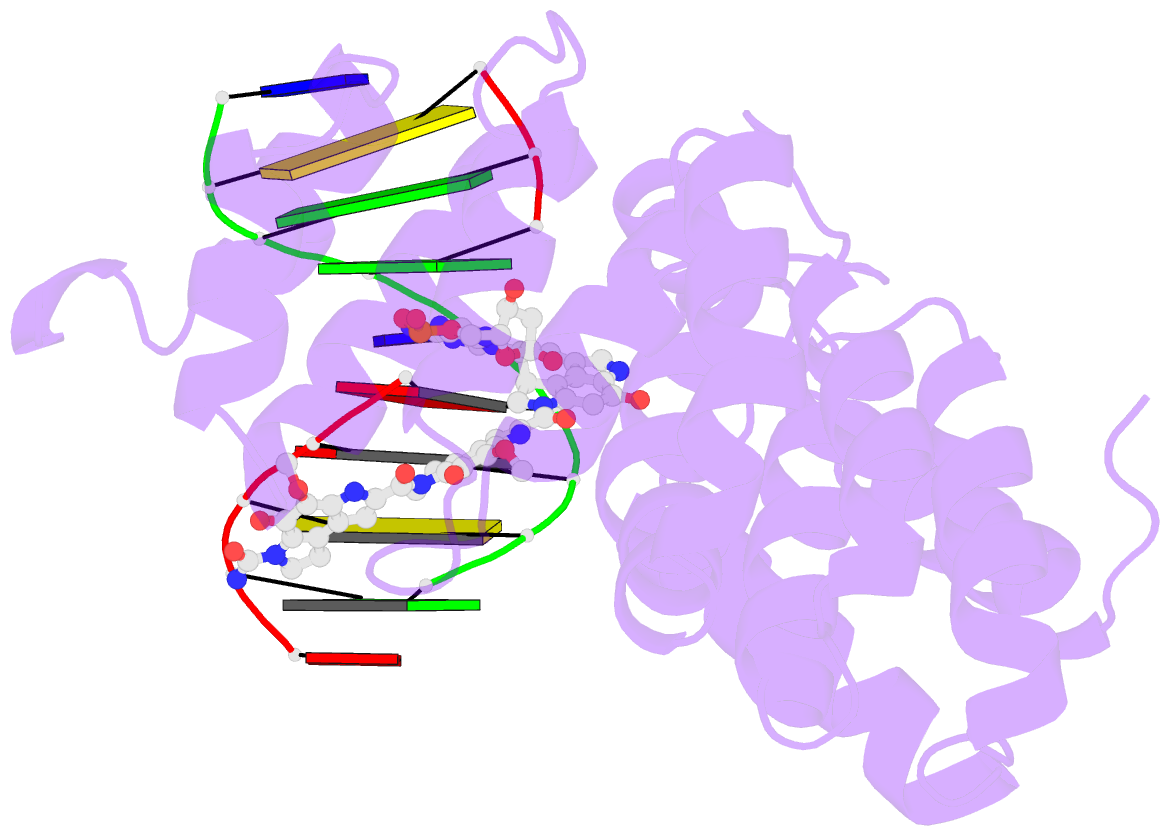
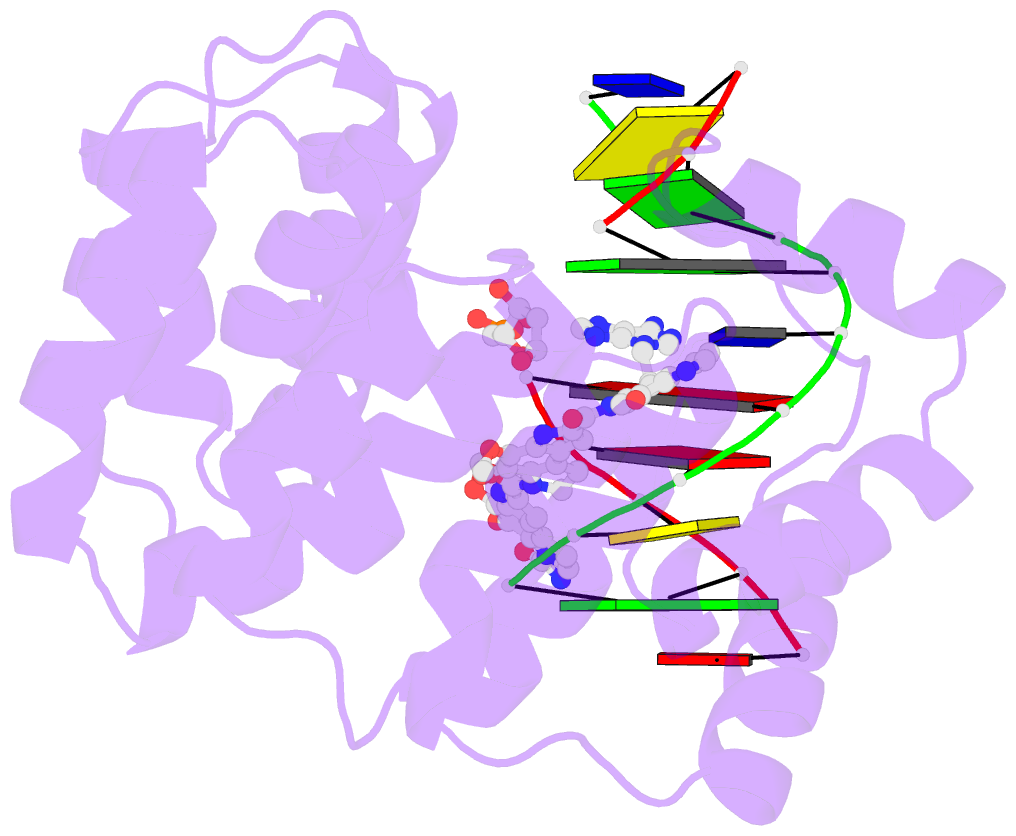
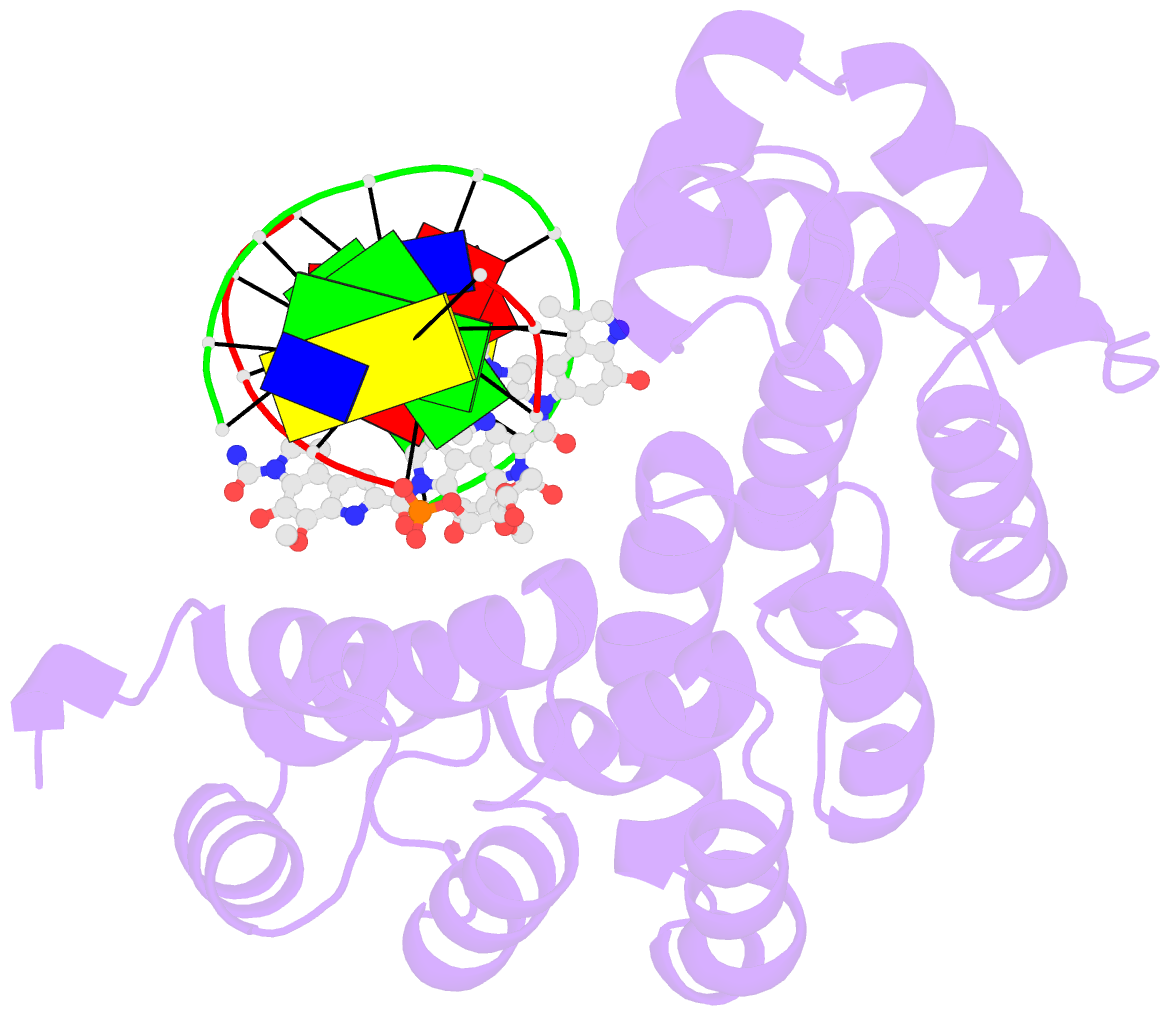
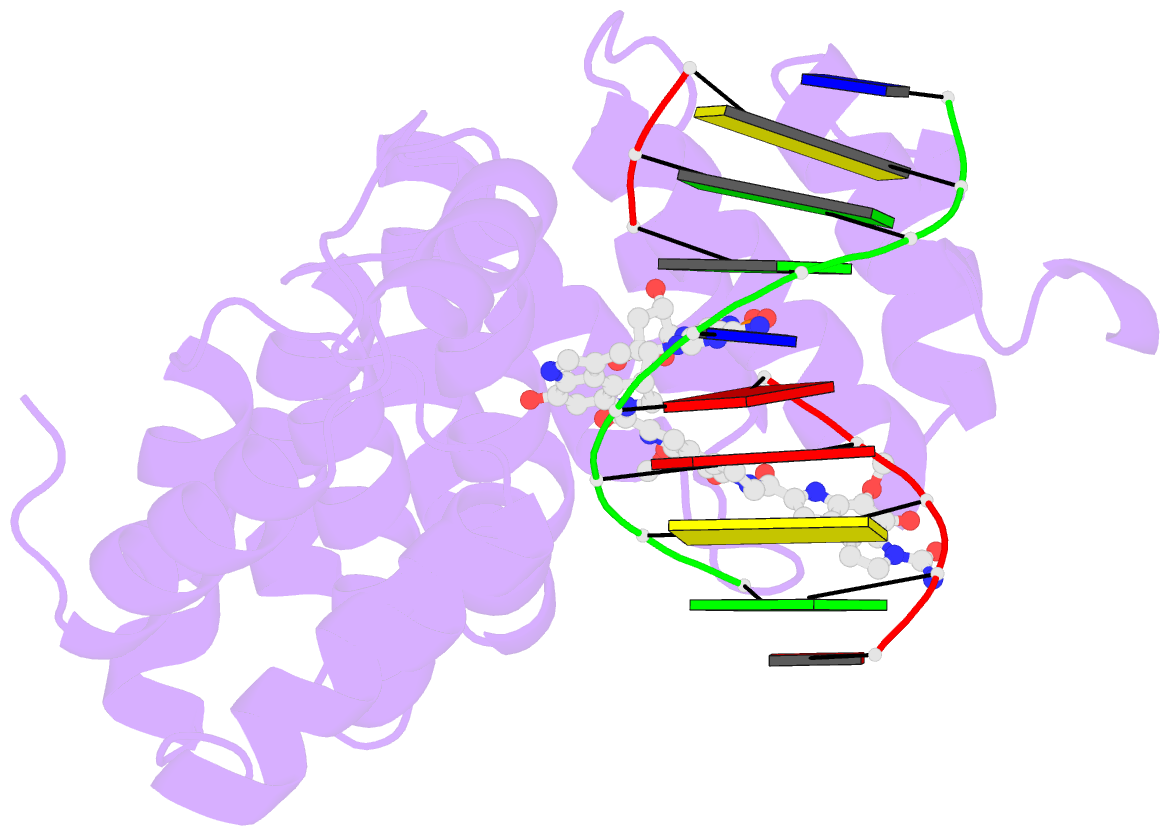
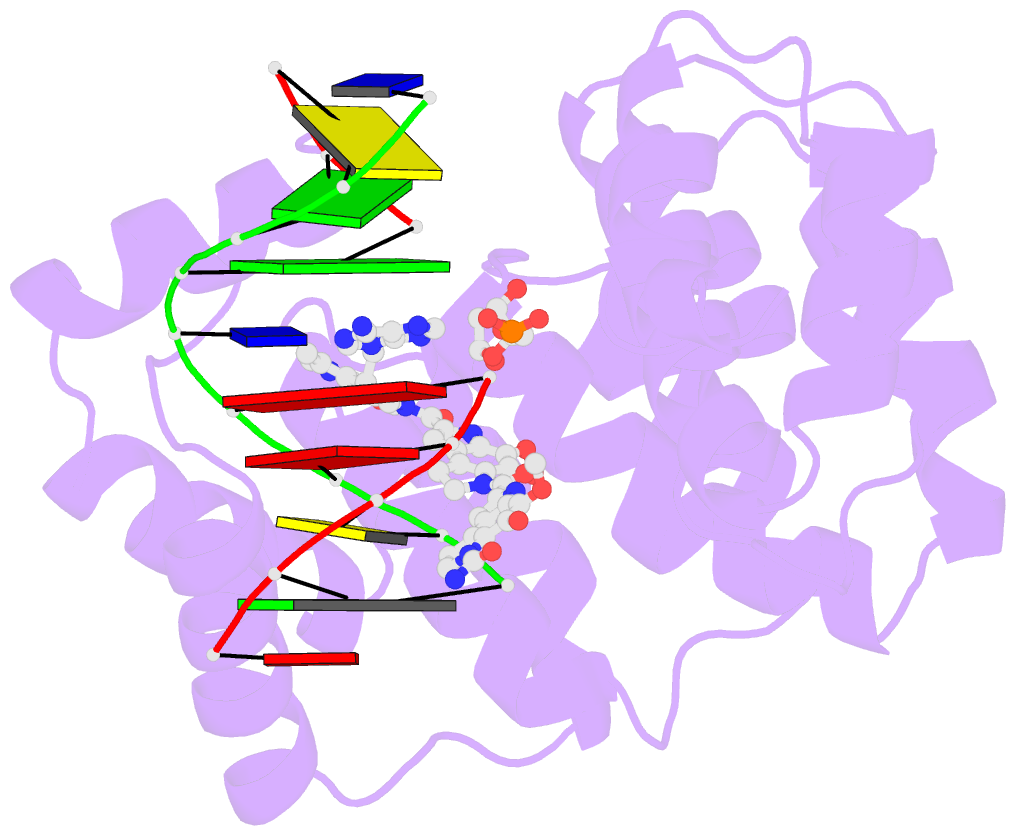
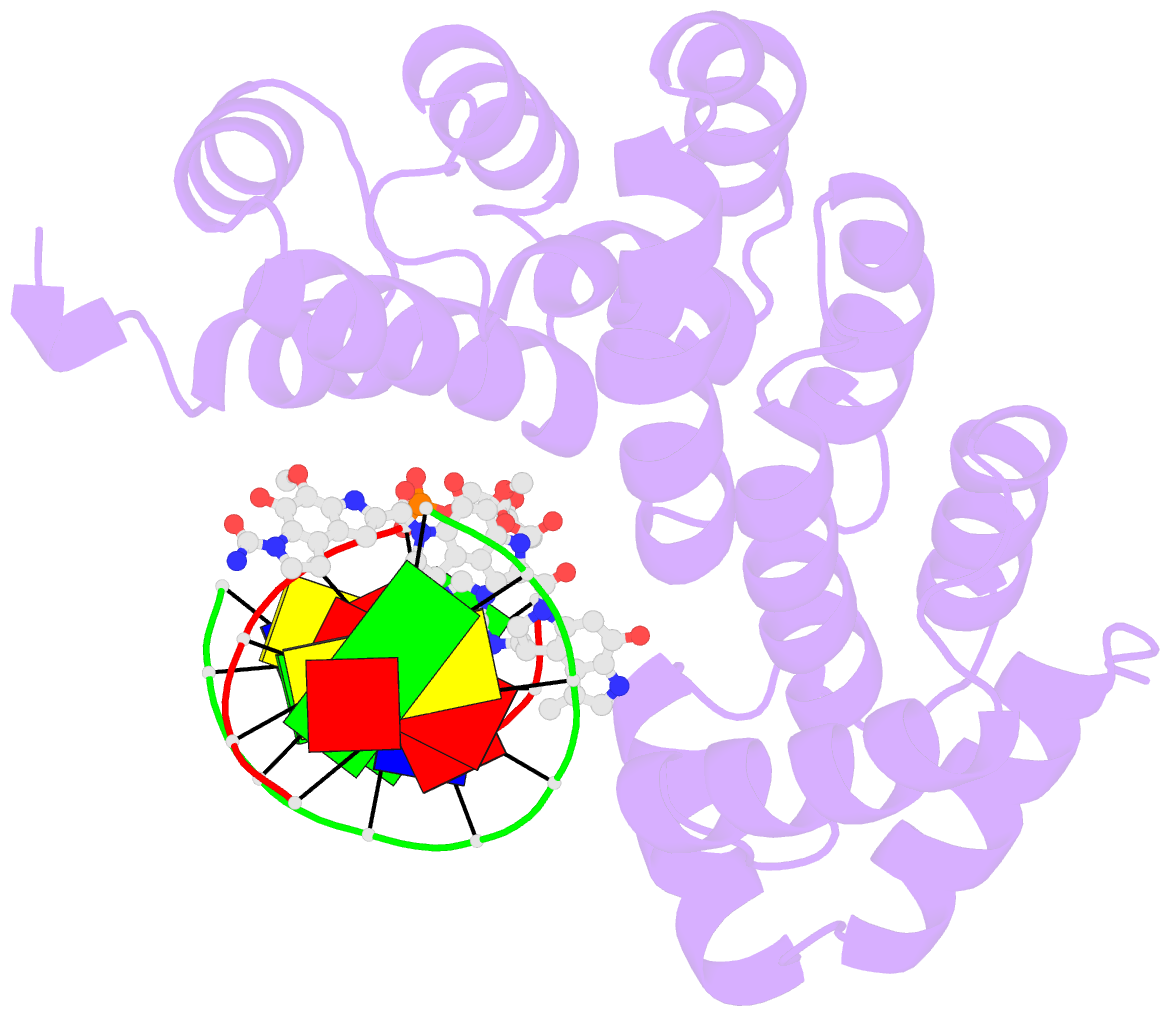
- Bacillus cereus DNA glycosylase alkd bound to a cc1065-adenine nucleobase adduct and DNA containing an abasic site
- Reference
- Mullins EA, Dorival J, Tang GL, Boger DL, Eichman BF (2021): "Structural evolution of a DNA repair self-resistance mechanism targeting genotoxic secondary metabolites." Nat Commun, 12, 6942. doi: 10.1038/s41467-021-27284-7.
- Abstract
- Microbes produce a broad spectrum of antibiotic natural products, including many DNA-damaging genotoxins. Among the most potent of these are DNA alkylating agents in the spirocyclopropylcyclohexadienone (SCPCHD) family, which includes the duocarmycins, CC-1065, gilvusmycin, and yatakemycin. The yatakemycin biosynthesis cluster in Streptomyces sp. TP-A0356 contains an AlkD-related DNA glycosylase, YtkR2, that serves as a self-resistance mechanism against yatakemycin toxicity. We previously reported that AlkD, which is not present in an SCPCHD producer, provides only limited resistance against yatakemycin. We now show that YtkR2 and C10R5, a previously uncharacterized homolog found in the CC-1065 biosynthetic gene cluster of Streptomyces zelensis, confer far greater resistance against their respective SCPCHD natural products. We identify a structural basis for substrate specificity across gene clusters and show a correlation between in vivo resistance and in vitro enzymatic activity indicating that reduced product affinity-not enhanced substrate recognition-is the evolutionary outcome of selective pressure to provide self-resistance against yatakemycin and CC-1065.