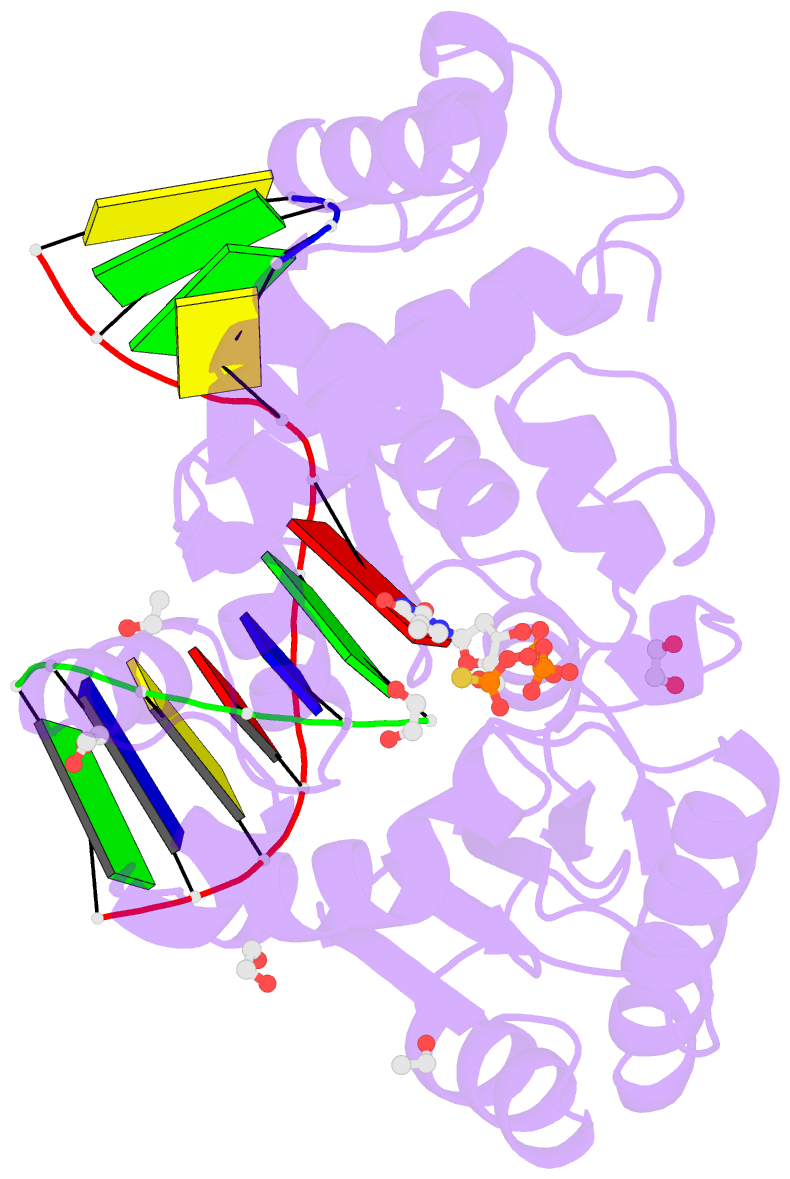
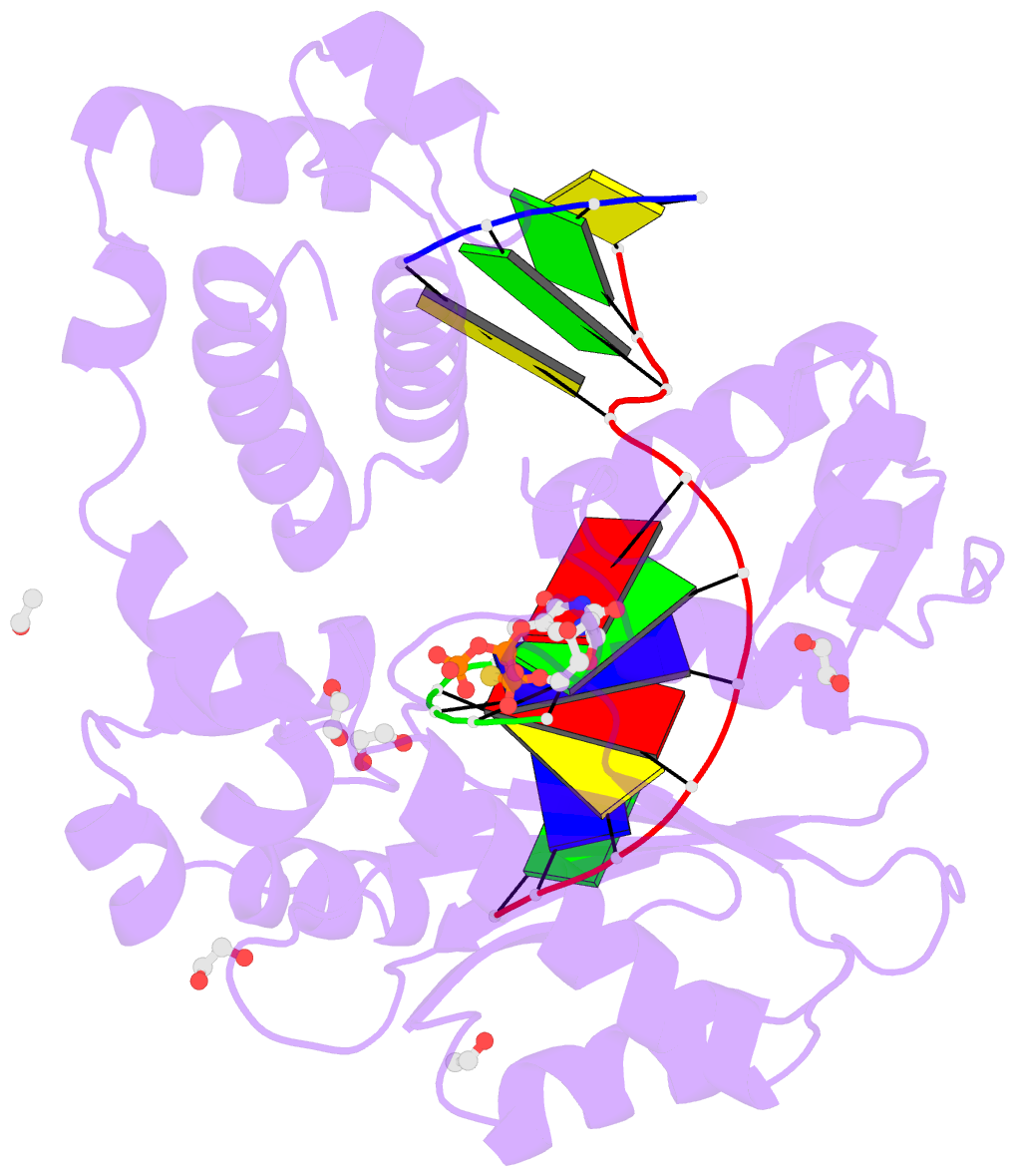
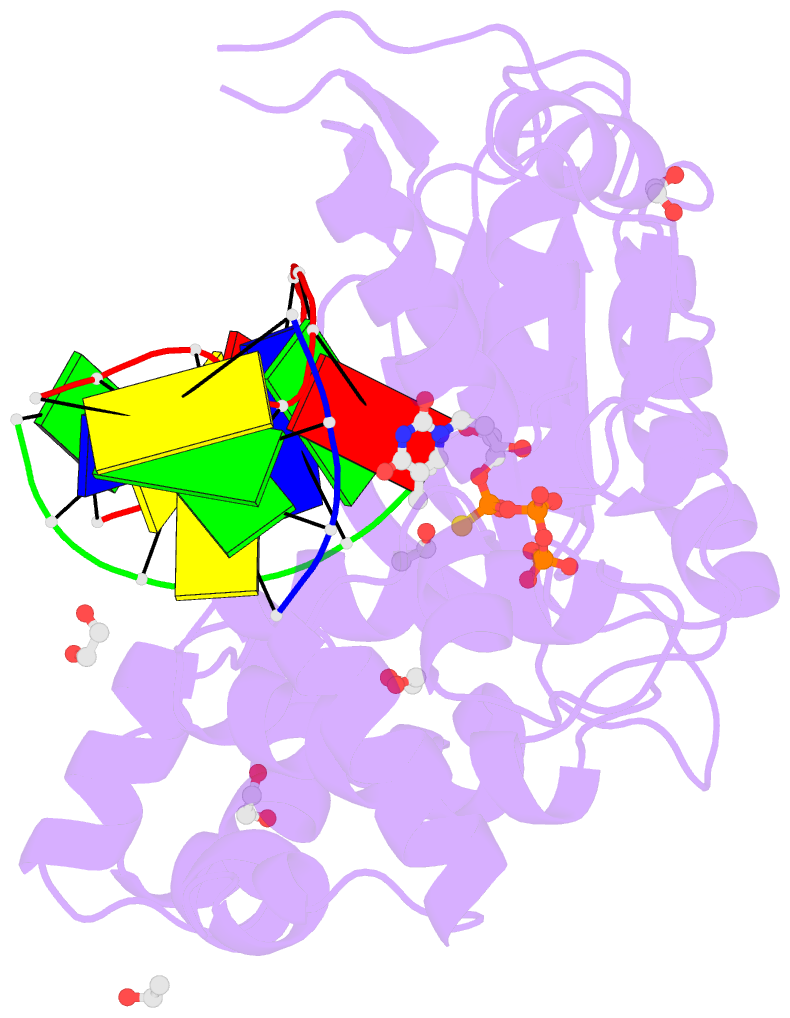
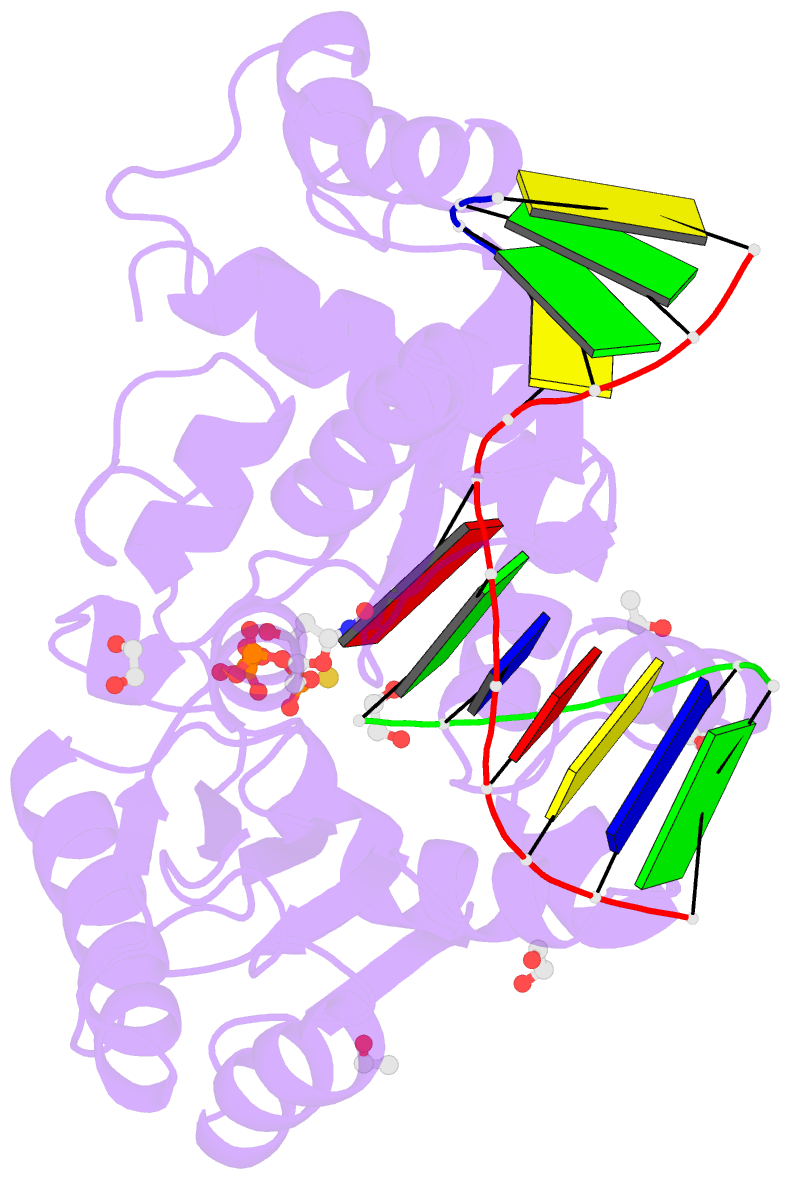
Summary information and primary citation
- PDB-id
- 7m4k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- X-ray (1.719 Å)
- Summary
- DNA polymerase lambda, ttpas:at ca2+ ground state ternary complex
- Reference
- Jamsen JA, Shock DD, Wilson SH (2022): "Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints." Nat Commun, 13, 3193. doi: 10.1038/s41467-022-30141-w.
- Abstract
- Efficient and accurate DNA synthesis is enabled by DNA polymerase fidelity checkpoints that promote insertion of the right instead of wrong nucleotide. Erroneous X-family polymerase (pol) λ nucleotide insertion leads to genomic instability in double strand break and base-excision repair. Here, time-lapse crystallography captures intermediate catalytic states of pol λ undergoing right and wrong natural nucleotide insertion. The revealed nucleotide sensing mechanism responds to base pair geometry through active site deformation to regulate global polymerase-substrate complex alignment in support of distinct optimal (right) or suboptimal (wrong) reaction pathways. An induced fit during wrong but not right insertion, and associated metal, substrate, side chain and pyrophosphate reaction dynamics modulated nucleotide insertion. A third active site metal hastened right but not wrong insertion and was not essential for DNA synthesis. The previously hidden fidelity checkpoints uncovered reveal fundamental strategies of polymerase DNA repair synthesis in genomic instability.